Figure 1.
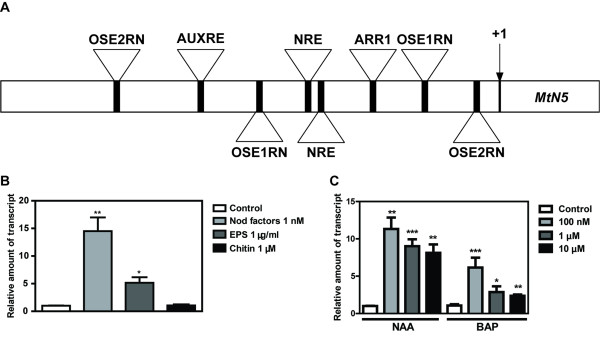
Bioinformatic analysis of the MtN5 promoter and expression of MtN5 in M. truncatula roots following treatments with bacteria-derived molecules and plant hormones. A. Schematic drawing of the putative MtN5 promoter and MtN5 open reading frame. The beginning of the ORF is identified by +1. The region analysed as the putative promoter encompasses 1.54 kb upstream the initiation codon. The following regulatory motifs are indicated: OSE1RN (OSE1ROOTNODULE; AAAGAT occurring in antisense orientation), OSE2RN (OSE2ROOTNODULE; AAGAG occurring in both sense and antisense orientations), NRE (AATTT), ARR1 (ARR1AT; GATT) and AUXRE (AUXREPSIAA4; GTCCCAT occurring in antisense orientation). B. M. truncatula roots were treated for 24 h with purified NFs (1 nM); for 48 h with EPS (1 μg/ml) and for 48 h with chitin (0.1 μM). C. M. truncatula roots were treated with α-naphtyl acetic acid (NAA) or with benzyl-amino-purine (BAP) at 100 nM, 1 μM and 10 μM for 48 h. The data were normalized to an internal actin control. The relative expression ratios were calculated using untreated roots as calibrator sample. The values reported are means ± SE (n=at least 3). Student’s t test was applied. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
