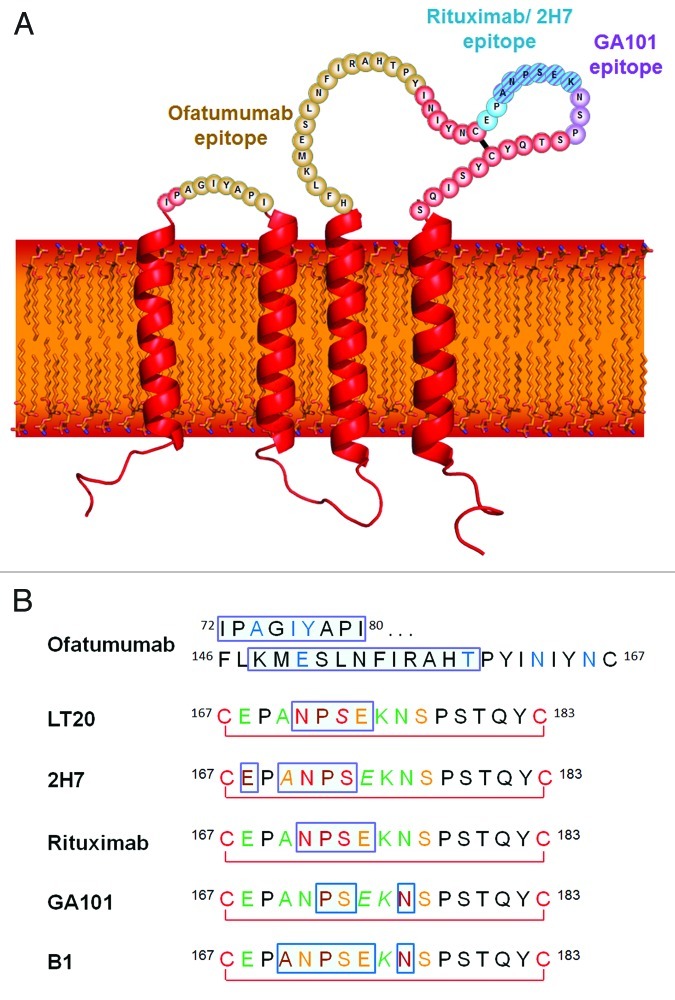
Figure 1. (A) The structure and topology of CD20 and the epitopes recognized by rituximab, ofatumumab and GA101. (B) Sequence alignment of CD20 epitopes recognized by CD20 antibodies based on published information. Core epitope residues are boxed in light blue. For 2F2 (ofatumumab), core epitope assignment is based on published work from Teeling et al. 46. For residues labeled in blue experimental evidence suggests a role in 2F2 binding. For the other antibodies, the following coloring scheme has been applied based on Pepscan results and FACS binding data of amino acid exchange mutants: green = almost any exchange tolerated at this position; brown = non-conservative exchange tested and not tolerated at this position; orange = conservative exchange tested and tolerated at this position; red = also conservative exchanges not tolerated at this position; black = position has not yet been evaluated. Italic font indicates that Pepscan and FACS binding results are discordant. Since the FACS binding results better reflect the native protein context, the coloring in such instants was based on the FACS binding data.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.