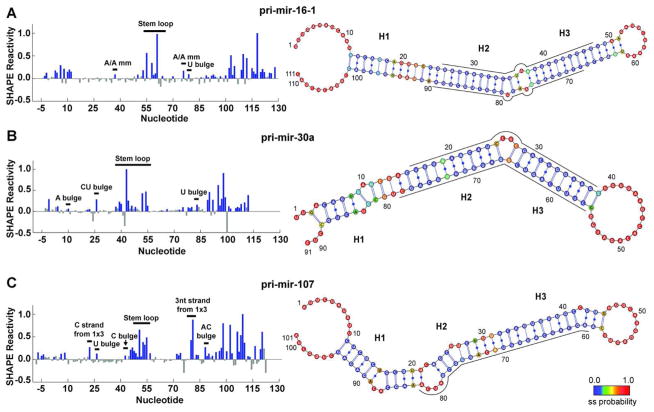
Figure 1.
SHAPE-constrained MC-Fold calculations yield secondary structures with embedded estimation of conformational dynamics. SHAPE reactivity traces (left) identify single-stranded nucleotides for (A) pri-mir-16-1, (B) pri-mir-30a, and (C) pri-mir-107. In the SHAPE reactivity traces, bar heights indicate the normalized mean reactivity constructed from 21 independent reactions. Blue filled bars indicate nucleotides with a positive mean reactivity and a magnitude greater than the uncertainty of the measurement. All grey bars indicate that the reactivity is either negative or has a mean magnitude below the uncertainty and are therefore considered insignificant. Addition of SHAPE-derived single-stranded constraints to MC-Fold calculations yields the combined probability of the nucleotide being single-stranded, which is mapped onto the most probable secondary structure (right). These probabilities are indicated in color as annotated in the color bar, ranging from most likely double-stranded (blue) to most likely single-stranded (red). Regions of high single-stranded probability divide the stems into three segments, labeled as H1, H2, and H3. Nucleotide numbering corresponds with the pri-miRNA numbering starting at 1 and the SHAPE cassette linkers (see Materials and Methods) being less than 1 and greater than the pri-miRNA length. Nucleotides corresponding to the mature miRNAs (as annotated in miRBase) are indicated by a line adjacent to the secondary structure diagram, which has been oriented such that the Drosha cut site is on the left in all three cases. Secondary. structure diagrams were generated in VARNA (54).