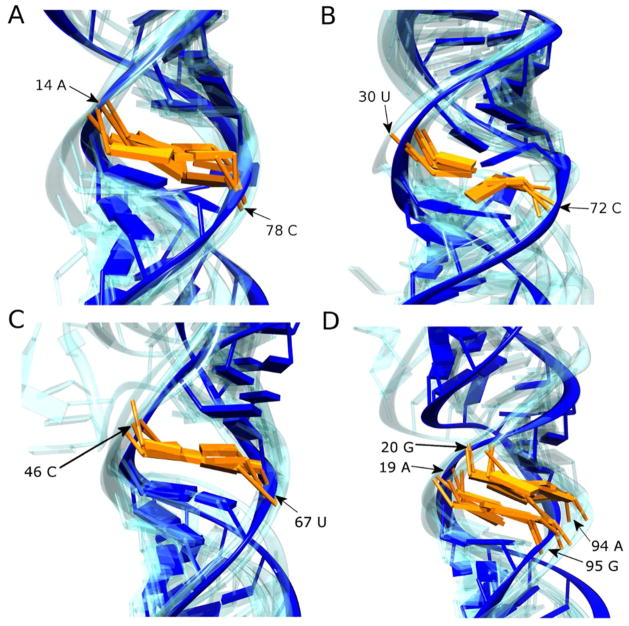
Figure 5.
The secondary structures of pri-miRNA molecules harbor multiple non-Watson-Crick mismatches that are predicted to be well-ordered by SHAPE reactivity. Expanded views of areas within the MC-Sym models representing these mismatches are shown for (A) A/C mismatch in pri-mir-30a, (B) U/C mismatch in pri-mir-107, (C) C/U mismatch in pri-mir-16-1, and (D) AG•GA internal loop in pri-mir-16-1. The nucleotides involved in the imperfections are colored orange (their position in the nucleotide sequence is also annotated) and the most probable structure is shown otherwise in solid blue, with the models from the other four members of the ensemble reported in Figure 3 shown in transparent blue. All models are aligned to the nearest stable Watson-Crick base-pair neighboring the imperfection in the most probable model.