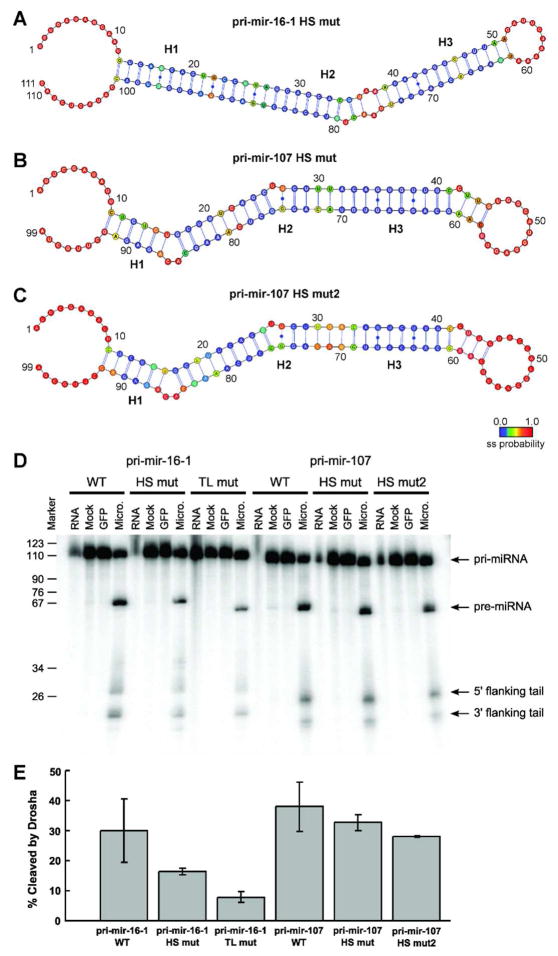
Figure 6.
Drosha processing of pri-miRNAs to pre-miRNAs in vitro confirms the necessity of hot spot flexibility for efficient cleavage. (A) The mutant pri-mir-16-1-HS has significantly reduced hot spot flexibility compared with wild-type, as established by combined SHAPE and MC-Fold analysis. (B) The mutant pri-mir-107-HS has the asymmetric 1-by-3 internal loop near the Drosha cleavage site replaced by a flexible C•C non-canonical base-pair, (C) while the mutant pri-mir-107-HS2 has only Watson-Crick base-pairs at the cleavage site, as established in both cases by combined SHAPE and MC-Fold analysis. (D) Denaturing gels for the processing of (from left to right) pri-mir-16-1 wild-type (WT), hot spot mutant (HS mut), and tetraloop mutant (TL mut) constructs; in addition to pri-mir-107 wild-type (WT), hot spot mutant (HS mut), and second hot spot mutant (HS mut2) constructs. In all six assays, lanes represent RNA collected prior to addition of Microprocessor (RNA), exposed to FLAG-beads with addition of cell lysate that did not express FLAG-tagged proteins for 5 minutes (Mock), exposed to GFP for 5 minutes (GFP), and 5 minutes after exposure to purified Microprocessor (Micro.). (E) The percentage of pri-miRNAs cleaved by the Microprocessor in vitro after 5 minutes averaged over three independent experiments. Cleavage is calculated as the sum of the intensities of the pre-miRNA product and the cleaved flanking tails divided by the sum of the intensities of the product, tails, and the remaining pri-miRNA substrate.