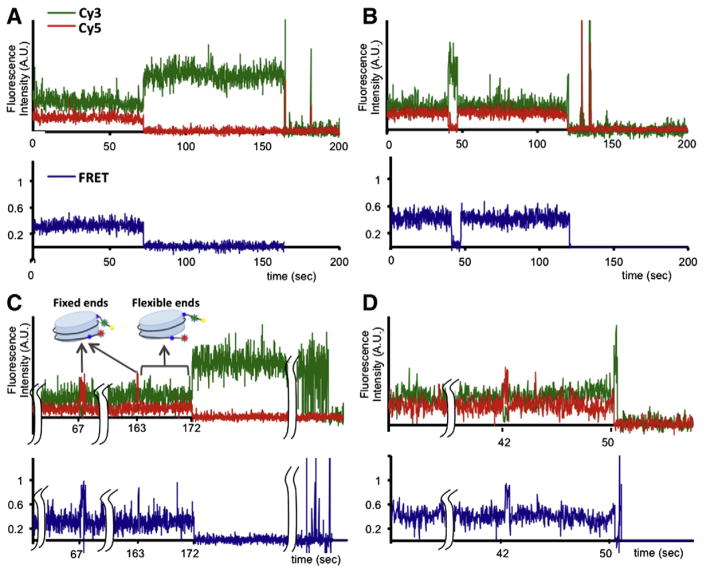
Fig. 4.
Effects of CpG methylation on the structure and structural dynamics of the terminal regions of nucleosomal DNA.5S rDNA nucleosomes display two unevenly distributed FRET states, a 0.29 FRET state (A) and a 0.4 FRET state (B) [14]. Signal integration time is 15 ms. After incubation with CpG methyltransferase, the population of nucleosome showing excursions to a high 0.74 FRET state was increased from 0.4% to 16% [14]. Examples of smFRET time trajectories that displayed excursions to a high 0.74 FRET state from 0.29 and 0.4 FRET states are shown in (C) and (D), respectively.