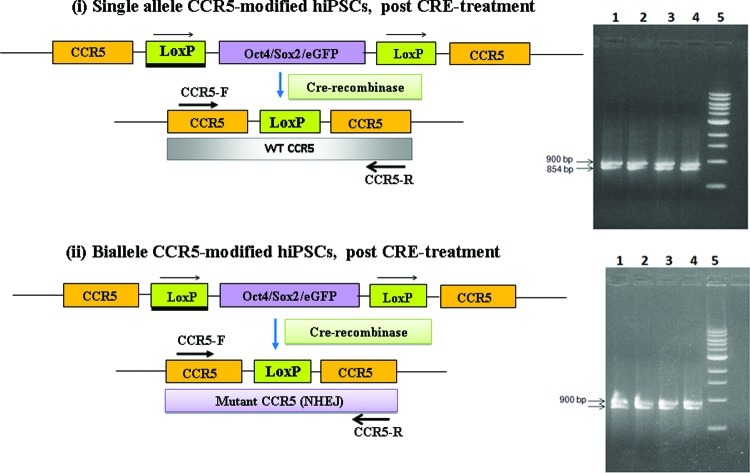
FIG. 4.
PCR and nucleotide sequence analysis of the mutated CCR5 locus of single-allele and biallele CCR5-modified hiPSCs. (i) Schematic diagram showing the configuration of the CCR5 chromosomal locus in single-allele CCR5-modified hiPSCs before and after Cre treatment. PCR amplification of the mutant CCR5 locus was performed using genomic DNA from single-allele CCR5-modified hiPSCs and primers flanking the CCR5-specific ZFN target sites. PCR analysis yields the expected 900 bp fragment for the CCR5 allele with a loxP site insertion and the expected 854 bp fragment for the CCR5 wild-type (WT) allele. (ii) Schematic diagram showing the configuration of the CCR5 locus in the biallele CCR5-modified hiPSCs before and after Cre treatment. PCR amplification of the mutant CCR5 loci was performed using genomic DNA from the 4 biallele CCR5-modified hiPSCs and primers flanking the mutation site(s). PCR analysis yields the expected 900 bp fragment for the CCR5 allele with a loxP site insertion and the expected 849, 856, 846 and 851 fragments, respectively, for the CCR5 allele mutated by non homologous end joining (lanes 1–4) for the CCR5-CRE-B-hiPSC1, CCR5-CRE-B-hiPSC2, CCR5-CRE-B-hiPSC3 and CCR5-CRE-B-hiPSC4 lines. The PCR fragments were then cloned and sequenced to determine the nucleotide sequence at the mutated CCR5 locus (see Table 1). Color images available online at www.liebertpub.com/scd