Table 1. Data-collection and refinement statistics for the RSK2–quercitrin complex.
Data from one native crystal were used for structure determination. Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 98.08, b = 40.69, c = 83.26, β = 114.3 |
| Resolution range (Å) | 30.00–1.80 (1.84–1.80) |
| No. of unique reflections | 28004 (1389) |
| R merge † | 0.117 (0.303) |
| 〈I/σ(I)〉 | 15.2 (4.65) |
| Completeness (%) | 99.8 (100) |
| Multiplicity | 4.3 (3.8) |
| Refinement | |
| Resolution range (Å) | 25.30–1.80 (1.84–1.80) |
| No. of reflections | 27492 |
| R work/R free ‡ | 0.17/0.20 |
| No. of atoms | |
| Protein (non-H) | 2266 |
| Inhibitor (all atoms) | 52 |
| Water | 260 |
| B factors (Å2) | |
| Protein | 37.8 |
| Inhibitor | 29.9 |
| Water | 45.4 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.98 |
R
merge = 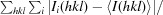
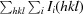 , where Ii(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of reflection hkl.
, where Ii(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of reflection hkl.
The crystallographic R factor R = 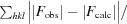
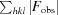 ; R
free =
; R
free = 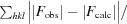
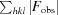 , where all reflections belong to a test set of randomly selected data.
, where all reflections belong to a test set of randomly selected data.
