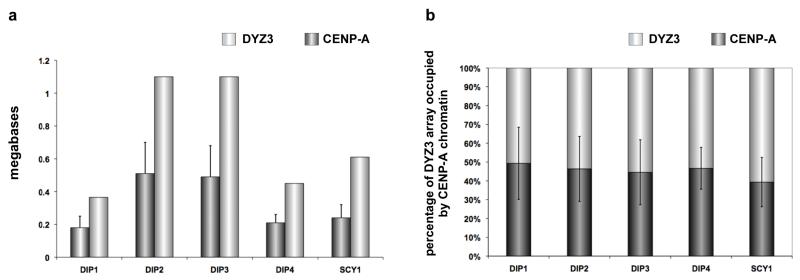
Figure 4. Human chromosome Y (HSAY) alpha satellite (DYZ3) array size compared to CENP-A chromatin domain size.
(a) DYZ3 arrays (light gray bars) were measured on different HSAYs present in human diploid (DIP) male lines or rodent-human somatic cell hybrids (SCY) in which HSAY was the only human chromosome. CENP-A chromatin domain size in megabases (dark gray bars) was determined using CENP-A immunostaining and FISH with a DYZ3 probe. As previously reported for DYZ3, arrays were distributed into two size groups: those <600kb and those >1Mb (Abruzzo et al., 1996). Accordingly, CENP-A domains clustered into groups that were either <200kb or >500kb. Error bars represent standard deviations in the genomic length (in megabases). (b) Calculation of the percentage of DYZ3 occupied by the CENP-A domain (dark gray bars) showed that the CENP-A domain was proportionally assembled on DYZ3 (light gray bars) that varied in size among different HSAYs. Typically, 45-50% of DYZ3 was occupied by CENP-A chromatin. Error bars represent standard deviations in the percentage of DYZ3 occupied by the CENP-A domain.