Abstract
ALG6-CDG (formerly named CDG-Ic) (phenotype OMIM 603147, genotype OMIM 604566), is caused by defective endoplasmic reticulum α-1,3-glucosyltransferase (E.C 2.4.1.267) in the N-glycan assembly pathway (Grünewald et al. 2000). It is the second most frequent N-glycosylation disorder after PMM2-CDG; some 37 patients have been reported with 21 different ALG6 gene mutations (Haeuptle & Hennet 2009; Al-Owain 2010). We report on the clinical and biochemical findings of five novel Caucasian South African patients. The first patient had a severe neuro-gastrointestinal presentation. He was compound heterozygous for the known c.998C>T (p.A333V) mutation and the novel c.1338dupA (p.V447SfsX44) mutation. Four more patients, presenting with classical neurological involvement were identified and were compound heterozygous for the known c.257 + 5G>A splice mutation and the c.680G>A (p.G227E) missense mutation. The patients belong to a semi-isolated Caucasian community that may have originated from European pioneers who colonized South Africa in the seventeenth/eighteenth centuries.
Introduction
ALG6-CG (phenotype OMIM 603147, genotype OMIM 604566) is a genetic disorder in the assembly of N-glycans due to ER alpha-1,3-glucosyltransferase deficiency (E.C 2.4.1.267). It was first reported in 1998 (Burda et al. 1998; Körner et al. 1998) and about 37 patients have been described (Imbach et al. 1999, 2000; Grünewald et al. 2000; Hanefeld et al. 2000; Westphal et al. 2000a, b, 2003; de Lonlay et al. 2001; Schollen et al. 2002; Newell et al. 2003; Sun et al. 2005; Vuillaumier-Barrot 2005; Eklund et al. 2006; Haeuptle and Hennet 2009; Al-Owain et al. 2010). The clinical presentation is mainly characterized by feeding problems and a moderately pronounced neurological disorder with psychomotor retardation, hypotonia, epilepsy, and internal strabismus (Grünewald et al. 2000; Newell et al. 2003). A minority of patients show other symptoms, particularly intestinal (such as protein-losing enteropathy (PLE)) and liver involvement (Damen et al. 2004). Striking biochemical features are unusually low serum cholesterol; blood coagulation factor XI and anticoagulation factors antithrombin, protein C, and protein S; as well as variable hypoalbuminemia; low serum LDL-cholesterol; and endocrinological abnormalities (Körner et al. 1998; Grünewald et al. 2000; Hanefield et al. 2000; Newell et al. 2003; Sun et al. 2005; Westphal et al. 2000a, b). Table 1 summarizes clinical and molecular findings in ten ALG6-CDG patients reported since 1998.
Table 1.
Genetic and clinical description of ALG6-CDG patients published from 1998 to 2011
| Reported ALG6-CDG patients | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| ALG6 gene mutation | c.998C>T (Homozygous and heterozygous) | Paternal: c.924C>A and c.391T>C, Maternal: c.998C>T (Heterozygous) | c.895-897 delATA and c.IVS3 + G>A (Heterozygous) | IVS 3 + 5G>A and c.998C>T or c.680G>A (Heterozygous) | c.509G>T and 1330_1332delCTT (Heterozygous) | c.391T>C and c.998C>T (Heterozygous) or c.391TT>C (Homozygous) | IVS3 + 3_3insT and c.998C>T (Heterozygous) | IVS7 + 2T>G and 897-897-899delAAT (Heterozygous) | c.338G>A and 10–12 Mb del(1)(p31.2p32.3) (Heterozygous) | c.482A>G (Homozygous) |
| Amino acid change | p.A333V(or combined with other mutation) | p.S308A, p.Y131H and p.A333V | In-frame deletion (delI299) and in-frame skipping | Aberrant splicing and p.A333V or p.G227E | p.S170I and in-frame deletion (ΔL444) | p.Y131H and p.A333V or p.Y131H alone | In-frame skipping and p.A333V | Aberrant splicing and in-frame deletion (delI299) | p.R113H and a deletion described on gDNA level | p.Y161C |
| Neurological abnormalities | ||||||||||
| Microcephaly/macrocephaly | Not reported | – | – | + (microcephaly) | Not reported | – | – | – | + (macrocephaly) | – |
| Hypotonia | + | + | + | + | + | + | + | + | + | + |
| Strabismus (internal or external if mentioned) | + (internal) | Not reported | + | + | – | + | + | – | + | – |
| Psychomotor retardation (including speech delay) | + | + | + | + | + (normal speech development) | + | + | + | + | + |
| Epilepsy | + | + | – | + | + | + | + | + | Not reported | + |
| Hypokinesia/ tremors | −/+ | – | – | – | – | – | + (tremors) | + (tremors) | – | + (hypokinesia) |
| EEG | Mostly normal | Normal | Abnormal | Not reported | Not reported | Normal | Not reported | Not reported | Normal | Not reported |
| Brain imaging | Cortical atrophy | Delayed myelination, slight cortical atrophy | Thin corpus callosum, mild cerebellar atrophy , mild widening of CSF spaces and ventricular system | Minimal cortical atrophy | – | Agenesis of corpus callosum (Westphal et al. 2003), cerebellar dysfunction (Miller et al. 2011) | Delayed myelination, normal cerebellum | Papilledema, but MRI normalized with age | Hypoplastic corpus callosum and absent septum pellucidum, white matter abnormality | Widening of CSF spaces and ventricular system, normal cerebellum |
| Optic dysfunction and/or -atrophy | −/+ | + | + | + | + | + (Miller et al. 2011) | Not reported | + | – | + |
| Dysmorphism | Broad nasal bridge, prominent forehead and large ears | – | – | Anormal long and narrow distal limbs, inverted nipples (noted at 6.5 months) | – | Broad nasal bridge | – | Abnormal fingers and toes (distal phalangeal hypoplasia with shortened toes and fingers) | Facial and limb abnormalities, deep set eyes, broad nasal bridge, loose skin, unusual fat distribution and other dysmorphic features | Severe body and facial abnormalities, inverted nipples and broad nasal bridge |
| Gastrointestinal symptoms | – | Severe protein-losing enteropathy, gastroenteritis and diarrhea | – | Protein-losing enteropathy | – | Gastrointestinal disturbances | Peripheral edema and diarrhea, abdominal distention, protein-losing enteropathy | – | Mild hepatomegaly | – |
| Other symptoms | Poor feeding, recurrent infections | Hormonal abnormalities, e.g., cortisol deficiency | – | Poor feeding, apnea at the age of 4.5 months | – | Abnormal endocrinological function | Poor feeding (anorexia), vomiting, cardiac abnormalities, recurrent infection | Apnea, cyanosis, abnormal hair growth and distribution, ovaria and hormonal abnormalities | Poor feeding, cardiac abnormality: biventricular hypertrophy | Poor feeding, cardiomyopathy, brachycephaly, bilateral cryptorchidism, bilateral esotropia |
| Reference | Imbach et al. 1999 (Genotype) and Grünewald et al. 2000 (Phenotype), Körner et al. 1998 | Westpal et al. 2000 (Dec) | Hanefeld et al. 2000 (Phenotype) and Westphal et al. 2000a (Genotype) | Imbach et al. 2000 (genotype), Du Plessis et al. 2001 (phenotype) and Schollen et al. 2002 | De Lonlay et al. 2001 | Westphal et al. 2003 and Miller et al. 2011 | Newell et al. 2003 | Sun et al. 2005 | Eklund et al. 2006 | Al-Qwaine et al. 2010 |
– : Not present
+ : Present (severity not indicated)
Twenty-one different mutations have been described but the majority of patients have the c.998C>T (p.A333V) mutation, either in a compound heterozygous or homozygous state (Imbach et al. 1999, 2000; Grünewald et al. 2000; Hanefeld et al. 2000; Westphal et al. 2000a, b, 2003; de Lonlay et al. 2001; Schollen et al. 2002; Newell et al. 2003; Sun et al. 2005; Vuillaumier-Barrot 2005; Eklund et al. 2006; Haeuptle and Hennet 2009; Al-Owain et al. 2010). Haeuptle and Hennet (2009) reported that most of the mutations affect amino acids positioned within the 11 TM domains, which compromise the integrity of the protein structure and alter the properties responsible for the binding of dolichol-linked glycans.
Serum transferrin IEF profiles of ALG6-CDG patients show a type 1 CDG pattern. LLO (lipid-linked oligosaccharides) analysis of fibroblasts shows an accumulation of Man9GlcNAc2 (Imbach et al. 1999; Burda et al. 1998; Marklova and Albahri 2007). A few isolated CDG-ALG6 patients (e.g., patients with the homozygous c.391TT>C, p.Y131H mutation) presented with a normal transferrin and LLO profile (Westphal et al. 2003; Miller et al. 2011). The Laboratory for Inborn Errors of Metabolism started screening for CDG in 1998 and Du Plessis et al. (2001) reported on the first South African CDG patient. We report on five novel Caucasian patients from South Africa with previously described mutations and one novel mutation.
Patients and Methods
Patients
The ALG6 patients in this study formed part of the semi-isolated Caucasian society, which can be defined as a population which is not found in a particular area in South Africa, but share the same historical past of forced segregation. These families may have originated from European pioneers who colonized South Africa in the seventeenth/eighteenth centuries, but further genealogy studies are needed in the future.
Patient 1 was referred to the Potchefstroom Newborn Screening (NBS) Laboratory in South Africa. A follow-up metabolic screen and CDG assays were suggested. Samples of patient 2, 3, 4, and 5 (Table 2) were initially referred to the Laboratory for Inborn Errors of Metabolism in Potchefstroom, South Africa, for a full metabolic investigation. The parents of the patients were all non-consanguineous and the patients were all Caucasian South Africans. Patients 1, 2, and 3 had no family ties, but patients 4 and 5 were first cousins. The latter two patients were not related to patients 1, 2, and 3. Informed consent was given by the families for the CDG genotype studies and the group formed part of the EUROGLYCANET project.
Table 2.
Clinical presentation of South African patients
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | |
|---|---|---|---|---|---|
| Gender and age | Male, died at 5 months | Male, died at 3.1 years | Male, 3.1 years | Female, 6.9 years | Male, 3.9 years |
| Neurological abnormalities | |||||
| Microcephaly | – | + | – | + | + |
| Hypotonia | ++ | ++ | ++ | ++ | +++ |
| Strabismus (internal or external) | – | + (internal) | – | + (internal) | + (internal) |
| Psychomotor retardation (including speech delay) | – | +++ | +++ | +++ | +++ |
| Epilepsy | – | +++ | +++ | +++ | +++ |
| Hypokinesia/Tremors | – | – | Hypokinesia | Hypokinesia | Hypokinesia |
| EEG | Not performed | Abnormal | Abnormal | Abnormal | Abnormal |
| Brain imaging | Thin corpus collosum | Mild cerebral atrophy | Mild cerebral atrophy | Mild cerebral atrophy | Severe atrophy, small cerebellum |
| Optic dysfunction/atrophy | – | + | + | ++ | +++ |
| Dysmorphisms | Enlarged eyes, micrognathia, broad-nasal bridge; camptodactyly (observed in fingers) | Broad nasal bridge | Broad nasal bridge | Broad nasal bridge; inverted nipples | Broad nasal bridge; small feet and fat pads on finger tips |
| Gastrointestinal symptoms | Protein-losing enteropathy with severe edema and ascites | – | Neonatal constipation and hepatomegaly | – | – |
| Other symptoms | Poor feeding | Recurrent infections | Poor feeding; recurrent infections | Recurrent infections | Poor feeding; recurrent infections |
Patients 1 to 5 represent CDG cases in five separate families
– : Absent
+ : Mild
++: Moderate
+++: Severe
Blood/Serum Transferrin IEF
Transferrin IEF analysis was performed on blood cards, of all five patients in this study at the Laboratory for Inborn Errors of Metabolism in Potchefstroom, South Africa, according to the methodology of Du Plessis et al. (2001). The validated blood card approach ensured a suitable sample for the transferrin analysis in a country, such as South Africa, in which transport and viability of samples could be compromised. Additional serum was collected from the five patients, who had a type 1 transferrin pattern on the blood card IEF analysis, and referred to the Academic Medical Centre (AMC), Amsterdam for confirmation.
ALG6 Mutation Analysis
Genomic DNA was isolated from EDTA blood of the patients, using the “NucleoSpin blood genomic DNA purification kit” (Macherey-Nagel, Germany, Düren). The ALG6 mutation analysis was performed by direct gene sequencing on DNA specimens, at the Department of Human Genetics of the University of Leuven (Imbach et al. 2000; Schollen et al. 2002) (The GenBank accession number of the ALG6 gene used in our study was NM_013339.3). An SNP database comparison and multiple alignment of the ALG6 protein were done for the novel mutation in this study.
Results
Clinical and Biochemical Description of Patients
Clinical data are summarized in Table 2. Patient 1 had severe PLE and neurological involvement. The newborn screening (NBS) result of patient 1 was normal except for an abnormal IRT (immunoreactive trypsinogen) result ([IRT = 145 μg/L, Ref value for 2–7 days < 75 μg/L]). Cystic fibrosis was excluded and no other metabolic abnormalities were observed. A metabolic screening and CDG analysis were requested, after the patient presented with clinical deterioration.
Patients 2 to 5 did not undergo NBS. Specimens of patient 2, 3, and 5 were sent to the metabolic unit for full metabolic screening and a transferrin IEF CDG analysis was specifically requested from clinicians. A CDG analysis was performed on a blood card sample of patient 4, after the exclusion of other metabolic abnormalities. Biochemical information on patients 2, 3, 4, and 5 was limited. Patient 2 had normal liver and thyroid function. No biochemical parameters were available for patient 3. Patients 4 and 5 had recurrent episodes of severe infections. No chromosomal abnormalities were documented. Additional blood parameters, which might have been of interest (e.g., lipidogram, coagulation and endocrinological tests), were not requested for any of the five patients in this investigation.
Blood/Serum Transferrin IEF
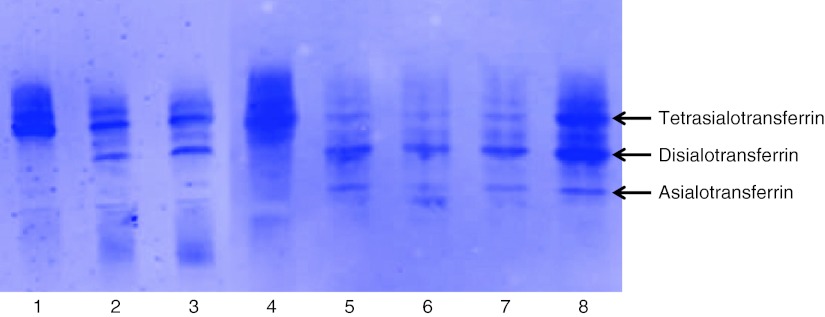
All five patients showed a type 1 serum transferrin IEF profile, that was most pronounced in patient 1 (almost no tetrasialotransferrin) (Fig. 1).
Fig. 1.
Lanes 1 and 4 represent control IEF transferrin patterns (the presence of tetratransferrin indicates a normal profile). Lane 2 indicates the IEF profile of patient 2 and lane 3 indicates the IEF profile of patient 4. Lanes 5, 6, and 7 represent the IEF profiles of patient 1. The IEF profiles were consecutively performed at 2 , 3 , and 4 months of age. Lane 8 represents the IEF profile of patient 3
ALG6 Mutational Analysis
Patient 1, with an intestinal-neurological phenotype, was found to be compound heterozygous for the c.998C>T (p.A333V) and c.1339dupA (p.V447SfsX44) mutations. Patient 2, 3, 4, and 5, with a predominantly neurological phenotype, were all compound heterozygous for the c.257 + 5G>A (originally described by Imbach et al. 2000 as the IVS3 + 5G→A mutation) and c.680G>A (p.G227E) mutations. The polymorphism, c.911C>T (p.S304F) was present in heterozygous state in patients 2, 3, 4, and 5. The carrier (father of patient 1) of the c.1339dupA (p.V447SfsX44) did not portray CDG-related symptoms.
The novel frameshift mutation, c.1339dupA (p.V447SfsX44), which we identified in this study, is not listed in the SNP database and affects a well-conserved amino acid in the ALG6 protein. Alignment studies were performed, using the computer software Alamut (version 1.5 rev. 32) of Interactive Biosoftware. The multiple alignment study of the ALG6 proteins of Homo sapiens (human), Pan troglodytes (chimpanzee), Rattus rattus (rat), Mus musculus (mouse), Canis familiaris (dog), Gallus gallus domesticus (chicken), and Tetraodon nigroviridis (pufferfish), indicated that the valine on position 447 is highly conserved. The frameshift mutation also results in the premature termination of the ALG6 protein (44 amino acids after the amino acid change) and therefore the formation of a truncated protein.
Discussion
The described patients showed two distinct phenotypes of ALG6-CDG deficiency. Patient 1 showed a severe, early-onset, neuro-gastrointestinal presentation comparable with previously described case studies (Westphal et al. 2000b; Damen et al. 2004; Newell et al. 2003). The patient was compound heterozygous for the prevalent c.998C>T (p.A333V) missense mutation (originally described by Imbach et al. 1999) and a novel c.1338dupA (p.V447SfsX44) frameshift mutation in the ALG6 gene. The p.A333V mutation is associated with a mild to moderate phenotype which indicates that the novel frameshift mutation (c.1338dupA), which results in the formation of a truncated ALG6 protein, probably explains the severe phenotype and early death of the patient as well as the unusually low serum tetrasialotransferrin. The phenotypic variation from the typical ALG6 deficiency concurs with previous literature of rare ALG6-CDG cases with multiple organ involvement and intestinal abnormality, e.g., PLE (Westphal et al. 2000b; Newell et al. 2003; Al-Owain et al. 2010).
Patients 2, 3, 4, and 5 presented with the classical ALG6-CDG phenotype comprising of a variable degree of neurological involvement, including hypotonia and resistant epilepsy. Other features in these patients were feeding problems, recurrent infections, and mild facial dysmorphism (Grünewald et al. 2000, Du Plessis et al. 2001). The c.257 + 5G>A splice site mutation (originally described by Imbach et al. 2000) in combination with the c.680G>A (p.G227E) missense mutation (originally described by Schollen et al. 2002) was identified in all four patients. The same combination of mutations in four patients may suggest that they have a common (most probably European) ancestor.
In summary, beside the patients with the classical neurological ALG6-CDG phenotype described in this study, we also identified an ALG6-CDG patient with an early onset of severe neurological and intestinal involvement associated with a novel frameshift mutation. This variation in ALG6 phenotype is in agreement with the literature summarized in Table 1 and suggests a genotype-phenotype correlation.
Acknowledgments
We thank the Laboratory for Inherited Metabolic Defects in South Africa (Potchefstroom) (http:///www.pliem.co.za) for the initial transferrin IEF analyses and for gathering of diagnostic material from South African CDG patients (a special thanks to Ansie Mienie, Elmarie Davoren, and Amaria van Huysteen). We appreciate sample referral from the Newborn Screening lab in South Africa (http://www.newbornscreening.co.za). We also want to express our gratitude to the colleagues of the Laboratory Genetic Metabolic diseases at the Academic Medical Centre, University of Amsterdam, who confirmed the South African CDG patients with serum transferrin IEF analyses. We thank Liesbeth Keldermans and Sandra Van Aerschot from the Department of Human Genetics and the Centre for Metabolic Diseases, at the University of Leuven, for excellent technical assistance. We are grateful to the patients and their families as well as to the referral doctors for their participation and support in this study.
Competing Interests
All parties confirmed that they have no competing interest for declaration.
Footnotes
Competing interests: none declared
References
- Al-Owain M, Mohammed S, Kaya N, Zagal A, Matthijs G, Jaeken J. A novel mutation and first report of dilated cardiomyopathy in ALG6-CDG (CDG-Ic): a case report. Orphanet J Rare Dis. 2010;5:1–4. doi: 10.1186/1750-1172-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Borsig L, de Rijk-van AJ, et al. A novel carbohydrate-deficient glycoprotein syndrome characterized by a deficiency of glycosylation of dolichol linked oligosaccharide. J Clin Invest. 1998;102:647–652. doi: 10.1172/JCI2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damen G, de Klerk H, Huijmans J, den Hollander J, Sinaasappel M. Gastrointestinal and other clinical manifestations in 17 children with congenital disorders of glycosylation type Ia, Ib, and Ic. J Pediatr Gastroenterol Nutr. 2004;38:282–287. doi: 10.1097/00005176-200403000-00010. [DOI] [PubMed] [Google Scholar]
- De Lonlay P, Seta N, Barrot S, et al. A broad spectrum of clinical presentations in congenital disorders of glycosylation: a series of 26 cases. J Med Genet. 2001;38:14–19. doi: 10.1136/jmg.38.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Plessis JA, Pretorius PJ, Lippert MM, et al. An atypical carbohydrate deficient glycoprotein patient in South Africa. S Afr Med J. 2001;91:392–394. [PubMed] [Google Scholar]
- Eklund EA, Sun L, Yang SP, Pasion RM, Thorland EC, Freeze HH. Congenital disorder of glycosylation 1c due to a de novo deletion and an hALG-6 mutation. Biochem Biophys Res Commun. 2006;339:755–760. doi: 10.1016/j.bbrc.2005.11.073. [DOI] [PubMed] [Google Scholar]
- Grünewald S, Imbach T, Huijben KM, et al. Clinical and biochemical characteristics of congenital disorder of glycosylation type Ic, the first recognized endoplasmic reticulum defect in N-glycan synthesis. Ann Neurol. 2000;47:776–781. doi: 10.1002/1531-8249(200006)47:6<776::AID-ANA10>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Haeuptle MA, Hennet T. Congenital disorders of glycosylation: an update on defects affecting the biosynthesis of dolichol-linked oligosaccharides. Hum Mutat. 2009;30:1628–1641. doi: 10.1002/humu.21126. [DOI] [PubMed] [Google Scholar]
- Hanefeld F, Körner C, Holzbach-Eberle U, von Figura K. Congenital disorder of glycosylation-Ic: case report and genetic defect. Neuropediatrics. 2000;31:60–62. doi: 10.1055/s-2000-7486. [DOI] [PubMed] [Google Scholar]
- Imbach T, Burda P, Kuhnert P, et al. A mutation in the human ortholog of the Saccharomyces cerevisiae ALG6 gene causes carbohydrate-deficient glycoprotein syndrome type-1c. Proc Natl Acad Sci. 1999;96:6982–6987. doi: 10.1073/pnas.96.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbach T, Grünewald S, Schenk B, et al. Multi-allelic origin of congenital disorder of glycosylation (CDG)-Ic. Hum Genet. 2000;106:538–545. doi: 10.1007/s004390050022. [DOI] [PubMed] [Google Scholar]
- Körner C, Knauer R, Holzbach U, Holzbach F, Lehle L, Von Figura K. Carbohydrate-deficient glycoprotein syndrome type V: deficiency of dolichol-P-Glc: Man9Glc-NAc2-PP-dolichyl glucosyltransferase. Proc Natl Acad Sci USA. 1998;95:13200–13205. doi: 10.1073/pnas.95.22.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklova E, Albahri Z. Screening and diagnosis of congenital disorders of glycosylation. Clin Chim Acta. 2007;385:6–20. doi: 10.1016/j.cca.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Miller BS, Freeze HH, Hoffmann SK. Pubertal development in ALG6 deficiency (congenital disorder of glycosylation type 1c) Mol Genet Metab. 2011;103:101–103. doi: 10.1016/j.ymgme.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell JW, Seo NS, Enns GM, McCracken M, Mantovani JF, Freeze HH. Congenital disorder of glycosylation 1c in patient of Indian origin. Mol Genet Metab. 2003;79:221–228. doi: 10.1016/S1096-7192(03)00089-1. [DOI] [PubMed] [Google Scholar]
- Schollen E, Martens K, Geuzens E, Gert Matthijs G. DHPLC analysis as a platform for molecular diagnosis of congenital disorders of glycosylation (CDG) Eur J Hum Genet. 2002;10:643–648. doi: 10.1038/sj.ejhg.5200858. [DOI] [PubMed] [Google Scholar]
- Sun L, Eklund EA, van Hove JLK, Freeze HH, Thomas JA. Clinical and molecular characterization of the first adult congenital disorder of glycosylation (CDG) Type Ic Patient. Am J Med Genet A. 2005;137:22–26. doi: 10.1002/ajmg.a.30831. [DOI] [PubMed] [Google Scholar]
- Vuillaumier-Barrot S. Molecular diagnosis of congenital disorders of glycosylation. Ann Biol Clin. 2005;63:135–143. [PubMed] [Google Scholar]
- Westphal V, Schottstädt C, Marquard T, Freeze HH. Analysis of multiple mutations in the hALG6 gene in a patient with congenital disorder of glycosylation 1c. Mol Genet Metab. 2000;70:219–223. doi: 10.1006/mgme.2000.3017. [DOI] [PubMed] [Google Scholar]
- Westphal V, Murch S, Kim S, et al. Reduced heparan sulfate accumulation in enterocytes contributes to the protein-losing enteropathy in a congenital disorder of glycosylation. Am J Pathol. 2000;157:1917–1925. doi: 10.1016/S0002-9440(10)64830-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal V, Xiao M, Kwok P, Freeze HH. Identification of a frequent variant in ALG6, the cause of congenital disorder of glycosylation-Ic. Hum Mutat. 2003;22:420–421. doi: 10.1002/humu.9195. [DOI] [PubMed] [Google Scholar]