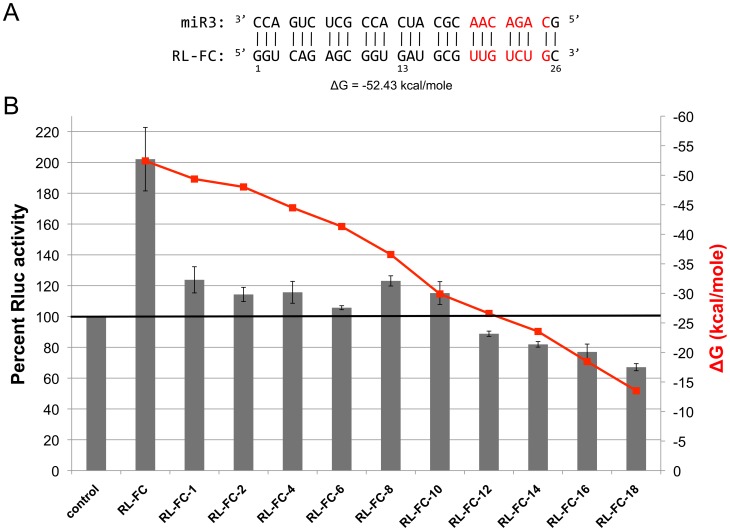
Figure 3. Analysis of mutations to a fully complementary (FC) miR3 target.
A) The numberings of nucleotides in FC with the seed sequence shown in red. The ΔG derived from base-pairings with miR3 is shown below. B) Mutations of RL-FC were carried out stepwise from the 5′-end of FC to reduce the base-pairings with miR3. Mutant transcripts were transfected into Giardia trophozoites along with miR3 and assayed for RLuc activity. The mean +/− SD from three independent experiments is presented. The ΔG values estimated from base-pairings with miR3 for each mutant is shown in an orange-colored line. The control column on the left represents the level of expression of each of the mutant transcripts in the absence of introduced miR3. They are each set at 100% and presented in a single column.