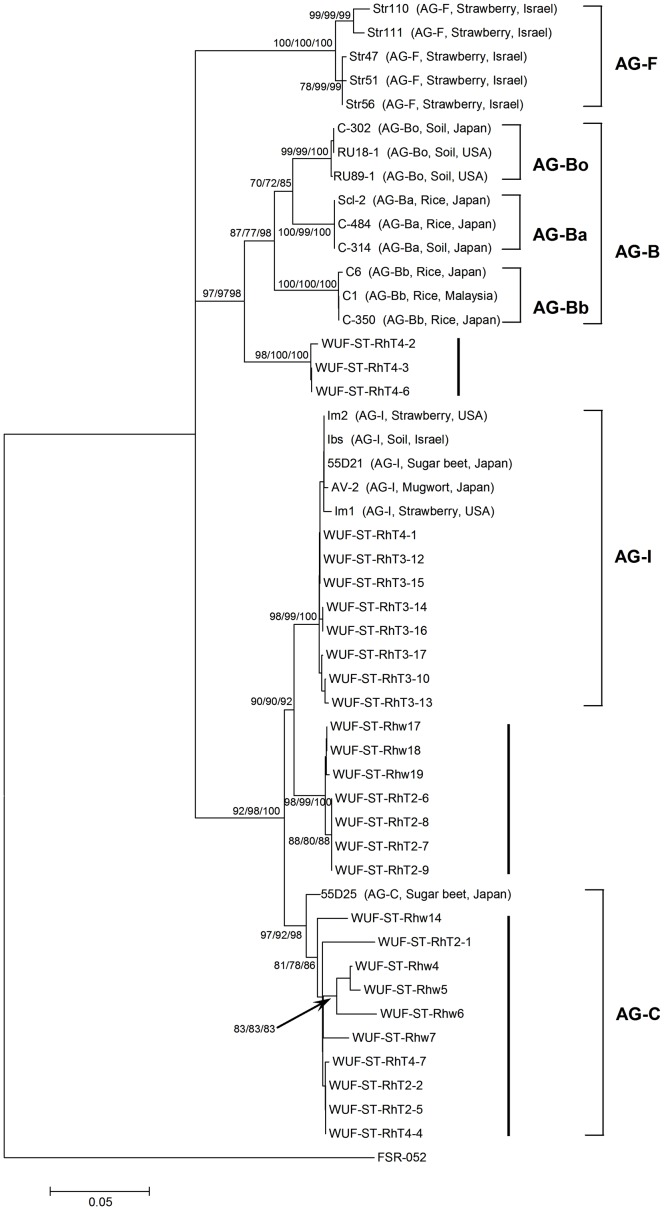
Figure 5. Phylogenetic relationships of the 28 pathogenic binucleate Rhizoctonia isolates in clade II ( Fig. 2 ) from this study with reference isolates from across the world based on the internal transcribed spacer sequences.
Trees were constructed using Maximum Likelihood (ML), Maximum Parsimony (MP) and Neighbor-Joining (NJ) analysis. All three methods generated the same tree topology. Tree branches were bootstrapped with 1,000 replications. Numbers at nodes indicate bootstrap values from ML/MP/NJ analysis, respectively. Only bootstrap values ≥70% are shown. Scale bar represents a genetic distance of 0.05 for horizontal branch lengths. The tree is rooted with isolate Athelia rolfsii FSR-052 (GenBank Accession No. AY684917) as the outgroup. The reference isolates are shown as isolate name followed by anastomosis group (AG), host and geographic origin of the isolate in parenthesis. The 28 isolates from this study are shown as isolate name only.