Table 2. MixMAP results in GLGC for IBC array loci with evidence for single SNP LDL-C association.
| Position | Single SNP p-values | |||||||||
| Locus‡ | Chr | Start | Stop | Gene Name‡ | Single SNP*1 | MixMAP*2 | # of SNPs | Min | Median | Max |
| LDLRAP9 | 1 | 25568422 | 25749764 | RHCE | + | - | 4 | 3.10E-10 | 2.07E-05 | 1.98E-003 |
| 1 | 25762009 | 25767440 | LDLRAP1 | +a | - | 5 | 6.38E-05 | 3.17E-04 | 3.59E-002 | |
| PCSK9 | 1 | 55267951 | 55304223 | PCSK9 | + | + | 34 | 1.93E-28 | 1.22E-05 | 0.8714 |
| ANGPTL3 | 1 | 62704220 | 62822181 | DOCK7 | + | - | 3 | 2.16E-17 | 3.17E-17 | 8.62E-017 |
| 1 | 62827868 | 62845701 | ANGPTL3 | + | - | 6 | 1.43E-17 | 0.18 | 0.973 | |
| SORT1 | 1 | 109745416 | 109745601 | PSMA5 | + | - | 2 | 1.56E-12 | – | 8.78E-011 |
| 1 | 109590236 | 109623689 | CELSR2 | + | + | 23 | 9.70E−171 | 2.31E-29 | 0.7796 | |
| 1 | 109622442 | 109630036 | PSRC1 | + | + | 7 | 4.93E-164 | 1.44E-08 | 0.3477 | |
| 1 | 109633806 | 109650249 | MYBPHL | + | + | 10 | 7.89E-28 | 1.68E-12 | 0.07608 | |
| 1 | 109652649 | 109742656 | SORT1 | + | + | 28 | 1.63E-23 | 1.14E-12 | 0.8676 | |
| APOB | 2 | 21052397 | 21165196 | APOB | + | + | 49 | 4.48E-114 | 7.51E-18 | 0.9401 |
| ABCG5/8 | 2 | 43921795 | 43958429 | ABCG8 | + | + | 21 | 1.73E-47 | 0.001336 | 0.9203 |
| 2 | 43893343 | 43924284 | ABCG5 | - | + | 25 | 8.14E-08 | 0.07115 | 0.4809 | |
| RAB3GAP1† | 2 | 136262314 | 136307216 | LCT | - | + | 11 | 1.13E-05 | 5.36E-05 | 0.6756 |
| HMGCR | 5 | 74667257 | 74693036 | HMGCR | + | + | 11 | 5.12E-45 | 5.70E-13 | 0.7414 |
| 74711473 | 74793312 | COL4A3BP | + | + | 9 | 2.90E-35 | 2.07E-12 | 0.4902 | ||
| TIMD4† | 5 | 155681482 | 156120506 | SGCDc | - | + | 66 | 3.38E-07 | 0.113065 | 0.8402 |
| 5 | 156445860 | 156469146 | HAVCR2c | - | + | 11 | 0.003134 | 0.009268 | 0.5286 | |
| HFE | 6 | 26196869 | 26204727 | HFE | + | - | 8 | 6.07-10 | 5.92E-03 | 6.64E-001 |
| HLA | 6 | 32512043 | 32535726 | HLA-DRA | + | + | 13 | 7.28E-13 | 0.01082 | 0.7028 |
| 6 | 31469689 | 31498389 | MICA | - | + | 20 | 2.60E-06 | 0.011105 | 0.6733 | |
| 6 | 31644203 | 31652541 | LTA | - | + | 18 | 0.0002275 | 0.01893 | 0.9282 | |
| 6 | 32000620 | 32025519 | C2 | - | + | 14 | 3.97E-05 | 0.041215 | 0.4409 | |
| 6 | 32899566 | 32917826 | TAP2 | - | + | 25 | 4.81E-07 | 0.116 | 0.8873 | |
| LPA | 6 | 160810340 | 160838285 | LPAL2 | - | + | 9 | 2.38E-06 | 0.0008496 | 0.6485 |
| 6 | 160873025 | 161011583 | LPA | + | + | 29 | 1.36E-15 | 1.36E-05 | 0.9738 | |
| NPC1L1 | 7 | 44519763 | 44551551 | NPC1L1 | + | + | 14 | 4.93E-11 | 7.35E-05 | 0.9729 |
| TRIB1 | 8 | 126506812 | 126573908 | TRIB1 | + | + | 49 | 2.83E-29 | 7.98E-10 | 0.9683 |
| ABO | 9 | 135197039 | 135229220 | SURF1 | + | - | 5 | 1.57E-12 | 1.60E-02 | 1.49E-001 |
| 9 | 135121293 | 135145180 | ABO | + | + | 15 | 4.60E-21 | 0.0003062 | 0.01966 | |
| ABCA1 | 9 | 106585724 | 107556417 | ABCA1 | +b | + | 121 | 1.12E-07 | 0.2582 | 0.979 |
| FADS1-2-3 | 11 | 61305135 | 61361791 | FADS1 | + | + | 9 | 1.75E-21 | 8.41E-21 | 0.0006352 |
| 11 | 61353788 | 61389758 | FADS2 | + | - | 6 | 2.12E-20 | 5.75E-04 | 7.88E-001 | |
| 11 | 61398293 | 61420267 | FADS3 | + | - | 7 | 8.80E-10 | 3.72E-04 | 6.20E-001 | |
| APOA5-A4-C3-A1 | 11 | 116024949 | 116145447 | BUD13 | + | + | 6 | 4.21E-99 | 4.91E-06 | 0.05234 |
| 11 | 116152068 | 116168917 | ZNF259 | + | + | 9 | 1.47E-26 | 4.01E-09 | 0.8141 | |
| 11 | 116157417 | 116170289 | APOA5 | + | + | 6 | 2.32E-16 | 7.16E-09 | 0.04245 | |
| 11 | 116172547 | 116202948 | APOA4 | + | - | 8 | 1.97E-08 | 0.069 | 0.732 | |
| 11 | 116212611 | 116233487 | APOA1 | + | - | 12 | 1.18E-09 | 4.12E-03 | 0.839 | |
| 11 | 116230513 | 116233840 | APOC3 | - | - | 2 | 7.93E-03 | – | 0.665 | |
| BRAP† | 12 | 110368991 | 110368991 | SH2B3 | + | - | 1 | 1.73E-09 | – | – |
| 12 | 110714419 | 110731337 | ALDH2 | + | + | 7 | 5.42E-09 | 5.92E-06 | 0.4789 | |
| 12 | 110971201 | 110971201 | C12orf30 | + | - | 1 | 6.89E-09 | – | – | |
| HNF1A† | 12 | 119890027 | 119923981 | TCF1 | + | + | 13 | 3.61E-15 | 0.03823 | 0.437 |
| CETP | 16 | 55548996 | 55576893 | CETP | + | + | 44 | 1.64E-12 | 0.003372 | 0.942 |
| HPR | 16 | 70640155 | 70671503 | HPR | + | - | 6 | 1.75E-22 | 1.35E-05 | 2.84E-001 |
| LDLR | 19 | 10663792 | 10663792 | ILF3c | + | - | 1 | 2.01E-14 | – | – |
| 19 | 11024562 | 11024562 | SMARCA4 | + | - | 1 | 1.74E-25 | – | – | |
| 19 | 11063306 | 11103658 | LDLR | + | + | 28 | 4.28E-117 | 6.98E-10 | 0.5643 | |
| CILP2 | 19 | 19185443 | 19329924 | NCAN | + | + | 12 | 1.42E-19 | 0.06043 | 0.5713 |
| 19 | 19324032 | 19366087 | KIAA0892 | + | - | 5 | 1.78E-15 | 5.68E-04 | 3.58E-001 | |
| 19 | 19515117 | 19524850 | CILP2 | + | - | 5 | 5.99E-21 | 0.506 | 0.631 | |
| 19 | 19526643 | 19584215 | PBX4 | + | - | 7 | 2.52E-18 | 6.89E-02 | 4.81E-001 | |
| 19 | 19580840 | 19619190 | EDG4 | + | - | 7 | 1.02E-17 | 0.065 | 0.051 | |
| APOE-C1-C2 | 19 | 49929652 | 49944944 | BCL3 | + | + | 6 | 4.21E-99 | 4.91E-06 | 0.05234 |
| 19 | 50021054 | 50021054 | BCAM | + | - | 1 | 6.18E-63 | – | – | |
| 19 | 50043777 | 50074877 | PVRL2 | + | + | 16 | 5.11E-67 | 1.05E-08 | 0.6158 | |
| 19 | 50081014 | 50119488 | APOE | + | + | 8 | 3.76E-110 | 2.67E-07 | 0.9906 | |
| 19 | 50100676 | 50100676 | TOMM40 | + | - | 1 | 3.76E-110 | – | – | |
| 19 | 50139001 | 50145079 | APOC4 | + | - | 5 | 1.08E-72 | 6.08E-03 | 0.458 | |
| 19 | 50139018 | 50149020 | APOC2 | + | - | 7 | 2.56E-10 | 0.074 | 0.652 | |
| TOP1† | 20 | 39225477 | 39230879 | PLCG1 | + | - | 2 | 5.99E-15 | – | 0.764 |
For the 31827 SNPs in 2960 genes interrogated in Global Lipids Genetic Consortium (GLGC) summary data [1], MixMAP detects 36 genes in 21 of 26 loci detected by single SNP analysis in GLGC. Coverage of genes detected by both approaches is higher, while the median p-value is generally lower, than that of genes detected by single SNP analysis alone. ‡Genes were assigned to a locus identified in the GLGC [1] if SNPs in that gene were within 500 Kb of or had linkage disequilibrium (LD) 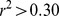 with the top SNP at a GLGC genome wide significant locus. †Genes designated at genome wide significant locus in GLGC were not directly interrogated in IBC array data;
with the top SNP at a GLGC genome wide significant locus. †Genes designated at genome wide significant locus in GLGC were not directly interrogated in IBC array data; 
 indicates corresponding gene detected and – indicates corresponding gene not detected;
indicates corresponding gene detected and – indicates corresponding gene not detected;  genome wide significant threshold (
genome wide significant threshold (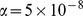 );
);  Bonferroni correction based on the number of genes (
Bonferroni correction based on the number of genes (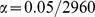 );
);  Gene is significant based on Single SNP approach in [1] (Table 1) but significant SNP is not included in IBC array under study;
Gene is significant based on Single SNP approach in [1] (Table 1) but significant SNP is not included in IBC array under study;  This gene is significant based on single SNP approach for TC and HDL in GLGC [1] (Table 1) and conditionally associated with LDL (Table 6 Supp);
This gene is significant based on single SNP approach for TC and HDL in GLGC [1] (Table 1) and conditionally associated with LDL (Table 6 Supp);  Limited LD (
Limited LD (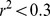 ) detected; however, assigned to same locus due to physical proximity (
) detected; however, assigned to same locus due to physical proximity (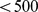 Kb) and/or multiple candidate genes in the region. These may represent signal for LDL-C independent of the established GLGC locus.
Kb) and/or multiple candidate genes in the region. These may represent signal for LDL-C independent of the established GLGC locus.
