Table 3. MixMAP in GLGC identifies IBC array loci that lack single SNP evidence for LDL-C association.
| Position | Single SNP p-values | Supporting | |||||||||
| Locus | Chr | Start | Stop | Gene Name | Single SNP*1 | MixMAP*2 | # of SNPs | Min | Median | Max | Evidence§ |
| PKN2 | 1 | 88918822 | 89073711 | PKN2 | - | + | 21 | 0.003906 | 0.06777 | 0.6081 | None |
| FN1 | 2 | 215938326 | 216066837 | FN1 | - | + | 44 | 1.56E-05 | 0.1254 | 0.9993 | †B [49], [50] |
| UGT1A1 | 2 | 234255355 | 234346384 | UGT1A1 | - | + | 20 | 3.53E-06 | 0.070225 | 0.9471 | A [18]–[20]; B [21] |
| PPARG | 3 | 12309416 | 12450557 | PPARG | - | + | 49 | 1.35E-05 | 0.1021 | 0.9917 | ?A,B,C[22]–[25] |
| DMDGH | 5 | 78331610 | 78402461 | DMGDH | - | + | 19 | 0.002283 | 0.08308 | 0.6427 | ‡A [51] |
| PPARD | 6 | 35419966 | 35503982 | PPARD | - | + | 45 | 0.002272 | 0.1943 | 0.8976 | A [31]–[34]; B [52] |
| CDK6 | 7 | 92074765 | 92304241 | CDK6 | - | + | 43 | 0.0006457 | 0.1083 | 0.8932 | None |
| VPS13B | 8 | 100143528 | 100936497 | VPS13B | - | + | 30 | 0.0003567 | 0.101375 | 0.861 | **A [42] |
| GAD2 | 10 | 26552050 | 26631994 | GAD2 | - | + | 22 | 0.006998 | 0.06269 | 0.3543 | None |
| GAB2 | 11 | 77604417 | 77802894 | GAB2 | - | + | 13 | 1.65E-05 | 0.0005164 | 0.9401 | None |
| APOH | 17 | 61629102 | 61657177 | APOH | - | + | 25 | 2.10E-05 | 0.1556 | 0.9846 | A [26]–[29] |
| NPC1 | 18 | 19366772 | 19415132 | NPC1 | - | + | 16 | 0.014 | 0.04201 | 0.3832 | A,B,C [35]–[38] |
For the 31827 SNPs in 2960 genes interrogated in Global Lipids Genetic Consortium (GLGC) summary data [1],  novel loci are detected by MixMAP alone. The median p-values for these genes tend to be lower than expected by chance. †No association with plasma lipids but mouse models support role in atherosclerosis; ‡No association with plasma lipids but cause of inborn error of choline metabolism; **No association with plasma lipids but implicated in Cohen syndrome in which truncal obesity is a feature; ?PPARG is a prominent gene for insulin resistance, type-2 diabetes mellitus, lipodystrophy and obesity; some data link PPARG to lipoprotein abnormalities;
novel loci are detected by MixMAP alone. The median p-values for these genes tend to be lower than expected by chance. †No association with plasma lipids but mouse models support role in atherosclerosis; ‡No association with plasma lipids but cause of inborn error of choline metabolism; **No association with plasma lipids but implicated in Cohen syndrome in which truncal obesity is a feature; ?PPARG is a prominent gene for insulin resistance, type-2 diabetes mellitus, lipodystrophy and obesity; some data link PPARG to lipoprotein abnormalities; 
 indicates corresponding gene detected and – indicates corresponding gene not detected;
indicates corresponding gene detected and – indicates corresponding gene not detected;  genome wide significant threshold (
genome wide significant threshold (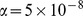 );
);  Bonferroni correction based on the number of genes (
Bonferroni correction based on the number of genes (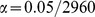 );
);  Based on published literature (see corresponding citations): A: human data; B: mouse/animal data; and C: cell biology.
Based on published literature (see corresponding citations): A: human data; B: mouse/animal data; and C: cell biology.
