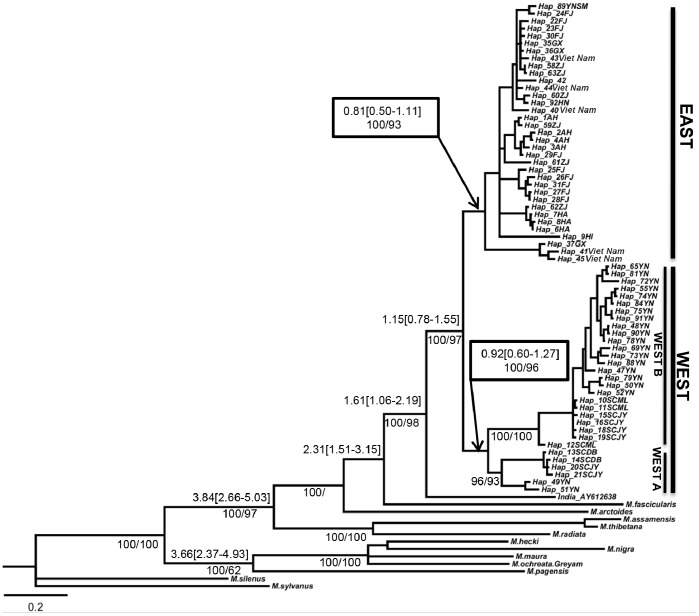
Figure 3. Bayesian inference matrilineal genealogy for the rhesus macaque, Macaca mulatta.
The genealogy is based on the concatenated sequences of three mtDNA gene sequences (16S+cytb+D-loop) consisting of 2260 bp. For the main nodes, Bayesian posterior probabilities (BPP≥90 retained) and bootstrap support from maximum parsimony (MP; 1000 replicates; MP≥50 retained) are shown above the branch, respectively. And the main nodes were dated by Bayesian inference using a Yule prior and the estimates are given below the branch, with 95% confidence intervals (CI).