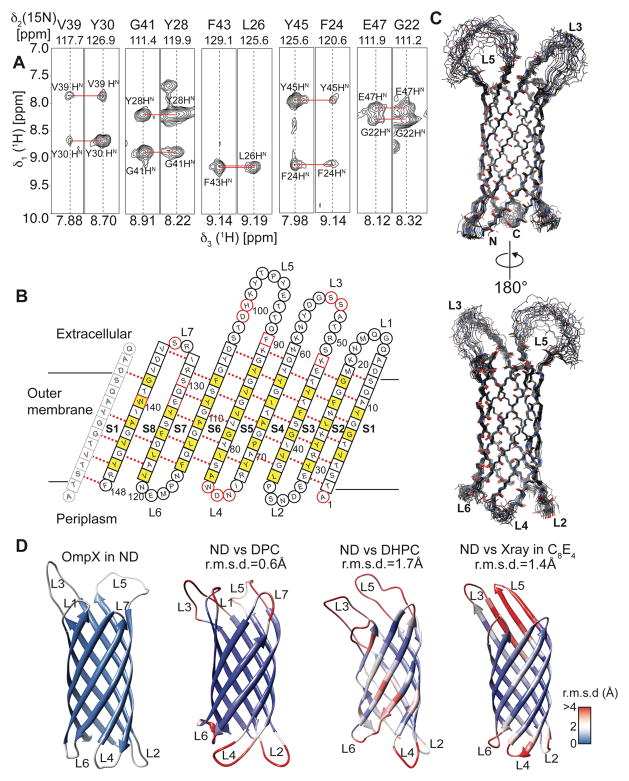
Figure 6.
Structure determination of OmpX in nanodiscs. (A) Example strips taken from a 3D-15N-edited-[1H,1H]-NOESY experiment showing long-range inter-β-strand contacts used for structure calculation. (B) Topology plot of OmpX. Residues facing towards the lipid bilayer are shown in yellow, unassigned residues in red. Residues in b-sheets are represented by squares whereas residues in loop regions are represented by circles. (C) The best-energy structural ensemble of (20 structures) of OmpX in nanodiscs has an r.m.s.d. of 0.32 Å between residues in β-sheets. (D) Comparison of the OmpX structure in nanodiscs (ND) with the NMR structures of OmpX in the detergents do-decylphosphocholine (DPC) (determined here), di-hexanoyl-glycero-phosphocholine (DHPC)15 and the crystal structure in n-octyltetraoxyethylene (C8E4)16. These structures are color-coded according to the r.m.s.d. values to individual residues of the nanodisc structure, as shown by the legend on the lower right. The r.m.s.d to ordered residues of the NMR structure in nanodiscs is indicated above each structure.