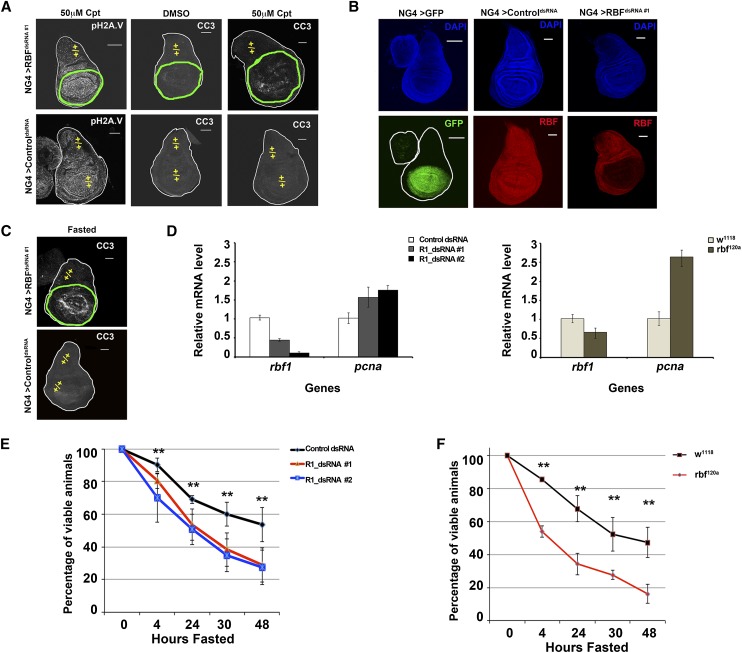
Figure 1.
Loss of the RBF1 protein leads to hypersensitivity to DNA damage and energetic stress. (A–C) Images shown are third instar wing discs. Bar, 100 μm. Genotypes are described in the Materials and Methods. The domain where the RBF1 protein has been depleted (green outline) or remains (yellow +/+) is designated. (A) Wing discs were treated with 50 μM Cpt or 50 μM DMSO as a control. DNA damage foci shown by pH2AV (grayscale). Apoptosis following Cpt challenge shown by CC3 (grayscale). (B) The nubbin-Gal4 expression domain is identified by the expression of a UAS-gfp (NG4 > GFP) (green). The RBF1 protein is efficiently depleted by “RBFdsRNA #1” (red), while ControldsRNA has no affect. Gross tissue architecture and like focal planes are indicated by DAPI (blue). (C) Fasting leads to apoptosis specifically in RBF1-depleted tissue, as indicated by CC3 (grayscale). (D) Error bars represent the standard deviation from the mean. mRNA levels were measured by quantitative real-time PCR (qPCR). RBF1 mRNA is efficiently depleted by both targeted dsRNAs. Depletion of RBF1 leads to increased levels of the traditional RBF1/dE2F target gene pcna. (E,F) Error bars represent the confidence interval (C.I.) of 95%. Mid-second instar larvae were fasted over 48 h. Reduced RBF1 protein leads to a statistically significant hypersensitivity to fasting when compared with appropriate control animals (at time points between 4 and 48 h the P-value < 0.01).