Abstract
The epithelial Na+ channel subunit-α (αENaC) of the distal nephron is essential for salt balance. We previously demonstrated that the histone methyltransferase Dot1a and its protein partner Af9 basally repress αENaC transcription in mouse inner medullary collecting duct type 3 (mIMCD3) cells and link aldosterone-elicited chromatin modifications to αENaC transcriptional activation. Af9 DNA-binding activity has never been demonstrated, and whether and where Af9 binds to the αENaC promoter to target Dot1a are unknown. The present study sought to identify functional Af9 cis-element(s) in the −57/+439 “R3” subregion of αENaC, the principal site for Dot1a-Af9 interaction, in mIMCD3 cells. We also exploited connecting tubule/collecting duct-specific Dot1l-deficient mice (Dot1lAC) to determine the impact of Dot1l inactivation on renal αENaC expression in vivo. mIMCD3 cell lines expressing αENaC promoter-reporter constructs harboring deletion of +74/+107 demonstrated greatly reduced association of Af9 and Dot1a by ChIP/qPCR. Aldosterone treatment resulted in further decrements in Af9 and Dot1a association with the αENaC promoter. Gel shift and antibody competition assays using wild-type and mutant oligomers revealed Af9-containing +78/+92 αENaC DNA-protein complexes in nuclear extracts of mIMCD3 cells. Mutation of the +78/+92 element resulted in higher basal αENaC promoter activity and impaired Dot1a-mediated inhibition in trans-repression assays. In agreement, mice with connecting tubule/collecting duct-specific knockout of Dot1l exhibited greater αENaC mRNA levels in kidney compared with control. Thus, we conclude that +78/+92 of αENaC represents the primary Af9 binding site involved in recruiting Dot1a to repress basal and aldosterone-sensitive αENaC transcription and that Dot1l inactivation promotes αENaC mRNA expression by eliminating Dot1a-mediated repression.
Keywords: chromatin, transcription, collecting duct, transcription factor, gene expression
the epithelial sodium channel (ENaC) plays a central role in collecting duct (CD) Na+ reabsorption, and hence the regulation of extracellular fluid volume and blood pressure (1). ENaC is expressed in the apical membrane of salt-absorbing epithelia of kidney, distal colon, and lung, where it constitutes the rate-limiting step in active Na+ and fluid absorption. ENaC consists of three subunits —α, β, and γ—encoded by the Scnn1a (referred to as “αENaC” in this report), Scnn1b, and Scnn1c genes. ENaC is also an important molecular target of aldosterone. In the CD, aldosterone administration or hyperaldosteronism induced by a low-Na+ diet increases αENaC gene transcription, without increasing β- or γ-subunit expression or αENaC mRNA turnover (14), and this response appears to be rate-limiting for ENaC activity in this segment. The physiological importance of αENaC to overall salt balance is highlighted by the finding that targeted inactivation of αENaC in the connecting tubule (CNT)/CD of mice results in severe renal salt wasting characteristic of a pseudohypoaldosteronism type I phenotype (5).
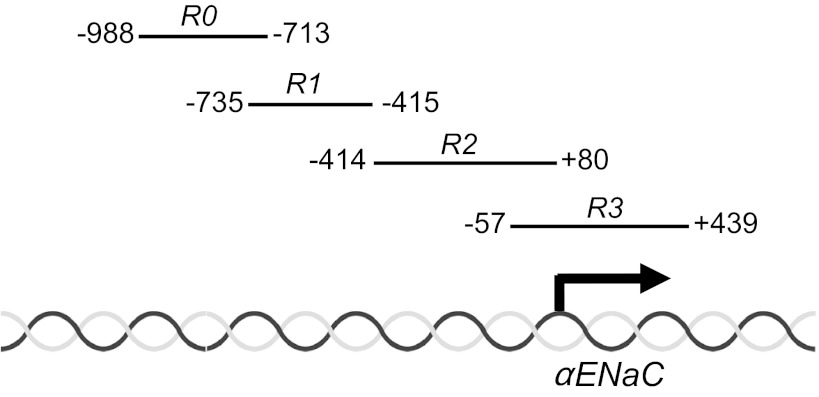
For a decade, it has been known that aldosterone stimulates αENaC transcription in CD cells (14). It had been assumed that this response was solely due to the action of aldosterone, liganded to the mineralocorticoid receptor (MR), acting at one or more hormone response elements in the αENaC promoter-enhancer. Indeed, promoter-reporter studies of the murine αENaC gene in CD cells revealed the functional importance of a glucocorticoid-responsive element (GRE) at −811 of the αENaC gene in the aldosterone response (12). However, mice with CNT/CD-specific knockout of the MR did not develop the severe salt-wasting phenotype (21) observed with targeted ablation of αENaC in these same segments (5), suggesting the importance of MR-independent pathways in αENaC regulation. Recently, though, we discovered an epigenetic pathway in mouse inner medullary collecting duct type 3 (mIMCD3) cells controlling a major component of basal and aldosterone-sensitive αENaC gene transcription, which involves combinatorial interactions of histone methyltransferase Dot1a with either the putative DNA-binding protein Af9 (26–28) or Sirt1 (25). Under basal conditions, Dot1a and Af9 are complexed with chromatin associated with four discrete subregions (which we termed R0-R3; Fig. 1) spanning −988 to +494 of αENaC (27). These associations facilitate the ability of Dot1a to hypermethylate Lys79 of histone H3, leading to a chromatin configuration that suppresses αENaC transcription (27). Sequential chromatin immunoprecipitation/qPCR (Re-ChIP/qPCR) studies determined that ∼75% of the basal Dot1a-Af9 association occurred at the −57/+494 “R3” subregion of αENaC, whereas roughly 15% occurred more distally at the R2 subregion, and only small but measurable amounts occurred at the R0 and R1 subregions (28). Moreover, Af9 appeared to be required for targeting Dot1a to the R3 subregion in the context of chromatin: overexpression of FLAG-Af9 resulted in Dot1a-Af9 association and hypermethylation of histone H3 Lys79 in sequential ChIP assays almost exclusively at the R3 subregion (28), and also resulted in repression of endogenous αENaC mRNA expression and the activity of αENaC promoter-luciferase constructs in mIMCD3 cells (27, 28). Conversely, RNA interference-mediated knockdown of Af9 caused the opposite effects (27).
Fig. 1.
Map of the subregions of the epithelial Na+ channel subunit-α (αENaC) 5′-flanking region. The nucleotide spans of the subregions R0-R3 are schematized.
We also demonstrated that aldosterone downregulates the expression of both Dot1a and Af9 (26) [as well as Sirt1 (25)], leading to decreased abundance of the Dot1a-Af9 repressor complex and histone H3 Lys79 hypomethylation at the αENaC promoter (26). Aldosterone also induces serum/glucocorticoid-regulated kinase 1 (Sgk1), which phosphorylates Ser435 of Af9, causing impairment of the protein-protein interactions of Dot1a with Af9 and dispersal of the Dot1a-Af9 complex from chromatin containing the αENaC promoter (28). This event results in hypomethylation of histone H3 Lys79 and release of transcriptional repression of the αENaC gene, contributing in large measure to the effects of aldosterone to increase αENaC gene transcription (28). This Sgk1 effect also occurs principally at the R3 subregion: in Re-ChIP/qPCR assays, mIMCD3 cells transfected with an Sgk1 expression construct exhibited an ∼80% reduction in Dot1a-Af9 occupancy and H3 Lys79 methylation at the R3 subregion of the αENaC gene (28). No changes in H3 Lys79 methylation were observed at the R2 subregion (the next highest site of Dot1a-Af9 binding under basal conditions) in these assays (28). Therefore, collectively our work to date indicates that the −57/+494 R3 subregion, which is well downstream of the −811 GRE, appears to be the principal site for Dot1a-Af9 binding and Dot1a-Af9-dependent epigenetic control of basal and aldosterone-induced αENaC gene transcription. It should be noted that we also demonstrated Dot1a-Sirt1 interactions that exert epigenetic control of basal αENaC gene transcription complementary to that of Dot1a-Af9 (25). The effect of Sirt1 to enhance Dot1a methyltransferase activity predominates at the R1 and R3 subregions (25).
Af9 was initially identified as one of the large number of fusion partners of the mixed lineage leukemia (MLL) gene that result from translocations at the 11q23 locus (11). Af9 expression in hematopoietic precursor cells promotes leukemic transformation (7), and the protein also appears to play important roles in anterior homeotic transformations during mouse development (6, 14), canonical Wnt signaling in mIMCD3 cells (9), and neurodevelopmental diseases, such as mental retardation, epilepsy, and ataxia in human patients (18). Af9 is comprised of conserved NH2- and COOH-terminal domains, centrally separated by a Pro/Ser-rich region that includes a nuclear localization signal. The NH2 terminus contains a YEATS domain, which is found in a variety of chromatin-modifying and transcription complexes. Recently, it was reported that erythroleukemic K562 cells transduced with Af9 showed significantly increased GATA1 promoter-reporter expression from the enhancer HS2 region, an effect that was lost if the YEATS domain was deleted from the Af9 construct (17). Aside from this report and results with artificial promoter-reporter constructs, very little is known about the function of Af9 and whether it truly functions as a sequence-specific transcription factor. ChIP analyses of the T-box brain protein 1 (Tbr1) gene suggested that Af9 associates near the Tbr1 transcriptional start site, promoting H3 Lys79 dimethylation of the region (presumably mediated by Dot1a), and interfering with RNA polymerase II binding to the region (2). However, Af9 DNA-binding activity and the recruitment and action of Dot1a were not directly demonstrated in that study. Indeed, potential cis-elements for Af9 binding in target genes have never been directly identified.
Chromatin contains genomic DNA compacted with histone and nonhistone proteins into a dynamic polymer. Basic residues in histones form ionic bonds to the acidic sugar backbone of DNA, allowing nonspecific interaction of the histone proteins with genomic DNA sequences. As an example, Dot1a is known to exhibit nonsequence-specific DNA binding activity in some chromatin contexts (15). In contrast, classical transcription factors bind directly to specific sequences in the DNA to affect changes in transcription. ChIP assays identify histone and nonhistone proteins associated with a DNA sequence in chromatin, and therefore may detect both sequence-specific and nonspecific protein binding to the DNA sequence. Moreover, these assays do not directly establish sequence-specific DNA-binding activity of a protein, since other protein partners in the cross-linked DNA-protein immunocomplex may direct DNA binding of the protein complex. Thus, our published ChIP results demonstrating Dot1a-Af9 associated with specific subregions of the αENaC promoter could be explained by one of three processes: 1) Dot1a, complexed with Af9, nonspecifically associates via a basic patch of histone H4 to the αENaC subregions studied in the ChIP assays; 2) Af9 acts as a transcription factor to directly bind the αENaC promoter in a sequence-specific manner, targeting its protein partner Dot1a to this location; or 3) Af9 serves as a bridging protein between Dot1a and an unknown transcription factor that anchors Dot1a-Af9 to a specific sequence of the αENaC promoter.
Recently, we described a new Dot1l conditional knockout mouse line (Dot1lf/f), which was generated using the Cre-LoxP system to allow inactivation of Dot1l methyltransferase and Af9-binding activities upon Cre-mediated recombination (3). This line was crossed with aquaporin 2 (Aqp2):Cre mice that express Cre recombinase driven by regulatory elements of the mouse Aqp2 gene (21). The resulting CNT/CD-specific Dot1l-deficient (Dot1lf/f Aqp2:Cre) mice were designated Dot1lAC (24). Compared with Dot1lf/f controls, Dot1lAC mice exhibit reciprocal changes in the number of principal cells (20% fewer) and intercalated cells (20% greater) (24). However, the important question of whether renal αENaC expression is upregulated in Dot1lAC mice, as our collective mIMCD3 cell data and Sgk1 knockout mouse data (28) would predict, was not examined.
Accordingly, the present study was designed to test whether Af9 directly binds to a discrete cis-element within the R3 subregion of the αENaC promoter, and if so, whether Af9 binding to its cis-element in this subregion serves as a platform for targeted and aldosterone-sensitive recruitment of Dot1a. We also compared the mRNA expression of ENaC genes in Dot1lAC vs. control mice to provide proof of concept that Dot1a basally represses αENaC expression in the CNT/CD. The results indicate that +78/+92 within R3 represents a functional Af9-binding site that plays a major role in recruiting Dot1a to the αENaC promoter for control of basal and aldosterone-sensitive αENaC transcription in mIMCD3 CD cells. Moreover, CNT/CD-specific Dot1l ablation resulted in upregulation of renal αENaC mRNA expression, presumably by relieving Dot1a-mediated repression of αENaC transcription.
MATERIALS AND METHODS
Reagents.
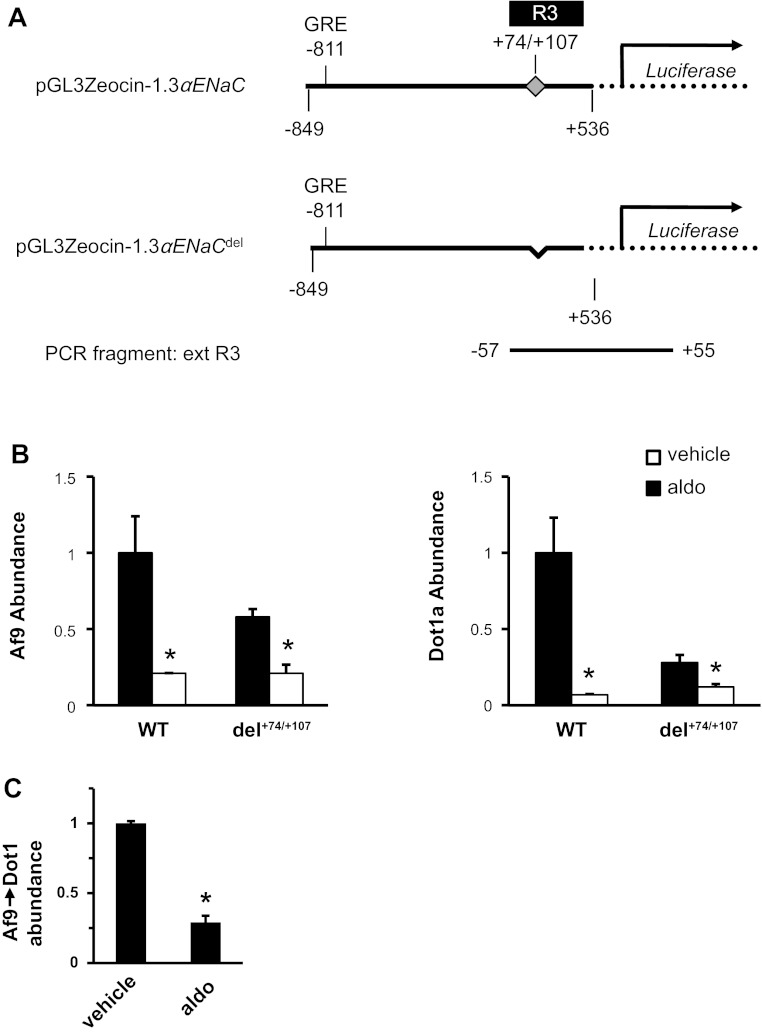
Aldosterone was from Sigma (St. Louis, MO). The Dual-Luciferase Reporter Assay System and pGL3-Basic were from Promega (Madison, WI). The SYBR GreenER qPCR SuperMix Universal and Lipofectamine 2000 reagent were purchased from Invitrogen (Carlsbad, CA). The Biotin 3′-End DNA labeling kit was from Pierce (Rockford, IL). The plasmids EGFP-Dot1a, FLAG-Af9, and pGL3Zeocin-1.3αENaC have been described (26–28). pGL3Zeocin-1.3αENaCdel, which lacks the native +74/+107 sequence in the αENaC promoter (see results and Fig. 2A), was generated by overlapping PCR using pGL3Zeocin-1.3αENaC as template. The 1.3-kb αENaC promoter and luciferase-coding cassette from pGL3Zeocin-1.3αENaC was also subcloned into the mammalian expression vector pcDNA3.1-Zeo to create pcDNA3.1-Zeo-1.3αENaC-Luc for use in trans-repression experiments. pcDNA3.1-Zeo-1.3αENaCΔ+78/+92-Luc harbors a cluster of point mutations between +78 and +92 (wild-type CAGGGAGCTGGAGG, mutant aAGGtctagtGAtt), which was introduced by the Quick Change Site-directed Mutagenesis Kit (Stratagene, Santa Clara, CA), according to the manufacturer's protocols. All mutations were verified by DNA sequencing analysis.
Fig. 2.
Chromatin immunoprecipitation (ChIP)/quantitative (q) PCR analysis of Af9 and Dot1a binding to +74/+107 in the αENaC promoter. A: diagram of the wild-type αENaC promoter-luciferase construct and its derivative harboring deletion of +74/+107. The position of the known glucocorticoid-responsive element (GRE) at −811 and the span of the R3 subregion are indicated. The PCR fragment “ext R3,” sharing the same 5′ start point as R3 (−57, relative to αENaC transcription site) and extending into the luciferase-coding region (+55, relative to luciferase translation start codon ATG), is shown at the bottom. B: ChIP/qPCR assay showing that aldosterone treatment results in decreased association of Af9 and Dot1a with the ext R3 region. Mouse inner medullary collecting duct type 3 (mIMCD3) cells carrying either the wild-type or del+74/+107 αENaC promoter-luciferase constructs were cultured in DMEM/F12 plus 10% charcoal-stripped serum and treated with vehicle or 1 μM aldosterone (aldo) for 2 h. The cells were then harvested and subject to ChIP assays with the antibodies specific for Af9 or Dot1a. Specific primers were used to amplify a 585- or 552-bp fragment, depending on the template derived from the wild-type or del+74/+107 αENaC promoter-luciferase constructs. The value for the final amplicon/input DNA obtained from the vehicle-treated cells harboring the wild-type promoter-reporter construct was set as 1, and the other values were normalized to it. Error bars indicate ± SE. *P value ≤ 0.05 vs. corresponding vehicle control; n = 3. C: Re-ChIP/qPCR assay showing aldosterone decreased Dot1-Af9 complex bound in the ext R3 region. Similar to B, chromatin from mIMCD3 cells transfected with the wild-type luciferase construct was sequentially immunoprecipitated with the Af9 antibody, followed by Dot1 antibody before qPCR. Error bars indicate ± SE. *P value ≤ 0.05 vs. corresponding vehicle control; n = 3.
Cell culture, aldosterone treatment, transient and stable transfections, and luciferase assays.
mIMCD3 cell culture, aldosterone treatment, transient and stable transfections, and luciferase assays of promoter activity were performed as described (26–28). Briefly, mIMCD3 cells were cultured at 37°C in a 5% CO2 environment in DMEM/F-12 plus 10% fetal bovine serum. Before 1 μM aldosterone or 0.01% ethanol was added as vehicle control at different time points, the cell medium was changed to DMEM/F-12 with 10% charcoal-stripped fetal bovine serum for at least 24 h. All cells were then harvested at the same time point. For stable transfections, mIMCD3 were transfected with pGL3Zeocin-1.3αENaC, pGL3Zeocin-1.3αENaCdel, pcDNA3.1-Zeo-1.3αENaC-Luc, or pcDNA3.1-Zeo-1.3αENaCΔ+78/+92-Luc, and clonal cell lines were established after Zeocin selection. Trans-repression experiments in mIMCD3 cells stably expressing pcDNA3.1-Zeo-1.3αENaC-Luc or pcDNA3.1-Zeo-1.3αENaCΔ+78/+92-Luc were performed by transient transfection of EGFP-Dot1a or insertless expression vector. Successful transfection was confirmed by demonstration of epitope-tagged proteins on immunoblots. Promoter activities were assayed using the Dual-Luciferase Reporter Assay System (Promega) and were normalized to cell protein content.
Gel shift and antibody competition assays.
Native or mutated +74/+107 double-strand oligomers were 3′ end-labeled with biotin using the Biotin 3′ End DNA labeling kit (Pierce). We centrally clustered the mutations between +78 and +92 within the mutant oligomer (wild-type CAGGGAGCTGGAGG, mutant aAGGtctagtGAtt, with mutations indicated by lower case letters), which were identical to the mutations introduced into pcDNA3.1-Zeo-1.3αENaCΔ+78/+92-Luc. Nuclear extracts were prepared from mIMCD3 cells and gel shift assays were conducted as detailed in our earlier work (25, 27, 28). For antibody competition assays, 0.2–2.0 μg of rabbit polyclonal antibodies against Af9 (Abcam, Cambridge, MA) or nonimmune IgG were preincubated with nuclear extracts for 20 min at 4°C, and then the biotin-labeled probes were added in the binding reactions and incubated at room temperature for 15 min before electrophoresis. After being transferred to nylon membranes, biotin-labeled DNA was detected by chemiluminescence using the LightShift Chemiluminescent EMSA kit (Pierce) according to the manufacturer's instructions.
ChIP/qPCR, Re-ChIP/qPCR, and RT-qPCR.
ChIP/qPCR and Re-ChIP/qPCR assays were performed and analyzed essentially as previously described (27, 28). For Re-ChIP, immunoprecipitates obtained with anti-Af9 were subjected to a second immunoprecipitation with antibody against Dot1a before qPCR. RT-qPCR to measure αENaC, βENaC, and γENaC mRNA levels in miMCD3 cells or mouse kidney utilized methods and primers as previously described (20).
Mutant mouse studies.
Generation of Dot1lAC mice and description of their polyuria phenotype on a normal-Na+ diet are detailed in our related manuscript (24). Briefly, we used a Dot1l conditional knockout line (Dot1lf/f) (3) and an Aqp2:Cre line (21) to inactivate Dot1l and thus abolish histone H3 Lys79 methylation in Aqp2-expressing cells, which are restrictively expressed in the CNT/CD. All experimental mice (2–5 mo old) were sex- and age-matched and had free access to water and a normal-Na+ (0.4%) diet. All animal studies were performed in accordance with National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by The University of Texas Health Science Center at Houston Animal Welfare Committee.
Data analysis.
Quantitative data are expressed as means ± SE and were analyzed for statistical significance by one-way ANOVA or Student's t-test as appropriate. P values ≤0.05 were taken as significant.
RESULTS
Dot1-Af9 basally associates with +74/+107 of the αENaC promoter and aldosterone decreases this association.
We previously demonstrated that Dot1a-Af9 complexes associate almost exclusively with the R3 subregion under basal conditions, elicit histone H3 K79 hypomethylation at this subregion, and activate the pGL3Zeocin-1.3αENaC promoter-reporter construct (26–28). Moreover, given the primacy of the R3 subregion in these effects, the minor occupation of Dot1a-Af9 in Re-ChIP/qPCR assays at the other αENaC promoter subregions (28), and the absence of sequence-specific Af9 binding on preliminary DNase I footprinting and gel shift assays of the separate R0, R1, and R2 subregions (data not shown), we elected to restrict our analysis of Af9 DNA-binding activity to the R3 subregion. To narrow down potential sequences involved in the Dot1a-Af9 binding within R3, we exploited comparative DNA sequence analysis of the proximal Tbr1 5′-flanking region, with which Af9 and hypermethlyated histone H3 Lys79 (suggesting association of Dot1a) have been shown to associate in ChIP assays (2). This analysis revealed that Tbr1 contains a region +36/+57 relative to the transcription start site that has 78% nucleotide identity to +78/+98 of αENaC. Accordingly, we restricted our initial structure-binding analysis to this sequence and neighboring nucleotides (+74/+107; the more extended sequence facilitated comparative analysis by ChIP assays) in R3 of αENaC.
We performed ChIP (immunoprecipitations with either anti-Af9 or anti-Dot1a) and Re-ChIP (immunoprecipitating first with anti-Af9 and then with anti-Dot1a) assays coupled with real-time qPCR to evaluate the significance of the +74/+107 sequence in mediating not only the basal and aldosterone-responsive association of Af9, but also the Af9-Dot1a complex with the αENaC promoter in mIMCD3 cells. We generated an αENaC promoter-luciferase construct identical to pGL3Zeocin-1.3αENaC except that +74/+107 was deleted, and designated it as pGL3Zeocin-1.3αENaCdel (Fig. 2A). We exploited the fact that this sequence is close to the luciferase-coding region contained in both constructs (Fig. 2A). This feature allowed us to couple the R3 forward primer (WZ770) with a reverse primer corresponding to aa 11–18 of the luciferase-coding region (WZ888), which would yield an extended R3 fragment (ext R3) of 585 or 552 bp, depending on whether the wild-type or +74/+107 deletion construct was the template (Fig. 2A). Importantly, this primer set does not amplify endogenous αENaC and so only detects the transfected constructs. In this way, Af9 and Dot1a association with wild-type and +74/+107 deletion constructs could be directly compared after stable transfection of mIMCD3 cells with either construct. For assays, the stable cell lines were cultured with medium containing 10% charcoal-stripped serum for 16 h and then treated with vehicle or 1 μM aldosterone for 2 h. We previously demonstrated that under the same culture conditions, aldosterone elicited histone H3 Lys79 hypomethylation and activation of the pGL3Zeocin-1.3αENaC promoter-reporter construct (26–28).
Focusing first on the differences in the ChIP/qPCR assays evident in the vehicle-treated cells (Fig. 2B), the cells transfected with pGL3Zeocin-1.3αENaCdel exhibited Af9- and Dot1a-binding levels of only 58 and 28%, respectively, of those observed in cells transfected with the wild-type promoter-reporter. This result indicates that, under basal conditions, +74/+107 is critical for a large component of Af9 binding, and >70% of Dot1a binding in the R3 subregion. Since Af9 binding was not totally abolished, this result also suggests the possibility that other sequences may exist within the R3 subregion to which Af9 directly or indirectly binds in the context of chromatin. In all cases, ChIP with the control IgG in parallel yielded only background signals and the products generally undetectable in agarose gel analysis (data not shown).
We next sought to determine whether +74/+107 was the principal sequence within the R3 subregion for aldosterone inhibition of Dot1a-Af9 complex abundance in the context of chromatin. As shown in Fig. 2B, aldosterone reduced Af9 binding in the ext R3 region from the cells carrying the wild-type reporter to 21 and 7%, respectively, compared with the vehicle-treated cells. Aldosterone lowered Dot1a binding from 28 to 12% in the cells carrying the mutant reporter. DNA immunoprecipitated by the Af9 antibody from the mIMCD3 cells carrying the wild-type promoter-reporter constructs was reimmunoprecipitated by the Dot1a antibody. As shown in the Re-ChIP/qPCR assay in Fig. 2C, aldosterone decreased association of the Dot1a-Af9 complex with the wild-type ext R3 to 29% compared with that in the vehicle-treated cells. Thus, +74/+107 accounts for >70% of the Dot1a-Af9 occupancy in the R3 subregion under basal conditions, and >70% of the regulatory dismissal of the complex from chromatin with aldosterone treatment.
Af9 directly binds the αENaC promoter.
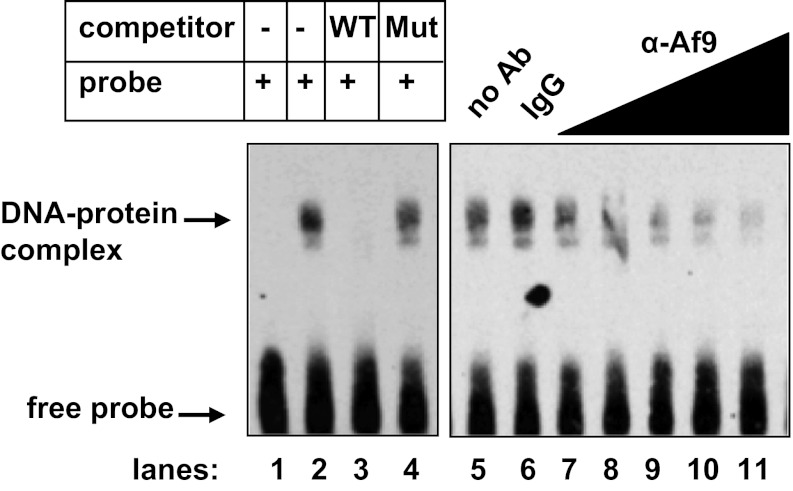
Given the ChIP/qPCR results demonstrating a critical role for +74/+107 in mediating association of Dot1a-Af9 complexes in chromatin with the αENaC promoter (Fig. 2B), we next sought to determine whether Af9 directly and specifically binds this sequence, and if it does, to narrow down the involved nucleotide sequence. Accordingly, we performed gel shift and antibody competition assays using biotin-labeled wild-type +74/+107 oligomers or +74/+107 oligomers harboring a cluster of 9 point mutations at +78/+92 and nuclear extracts harvested from mIMCD3 cells. A sequence-specific gel shift was consistently observed only with the wild-type +74/+107 oligomer (Fig. 3, lanes 1 and 2). The DNA-binding activity observed was sequence-specific, because binding was competed by a molar excess of unlabeled wild-type +74/+107 oligomer, but not by the mutant oligomer (Fig. 3, lanes 3 and 4). The fact that the mutant oligomer did not bind nuclear proteins or compete with the wild-type oligomer for binding indicates a requirement for +78/+92 in Af9 binding. Antibody competition analysis showed that nuclear extracts incubated with anti-Af9 antibody exhibited much less +74/+107 DNA-binding activity compared with controls incubated with IgG, but no supershift (Fig. 3, lanes 5–11). This result most likely indicates that the Af9 antibody disrupted the Af9-DNA interaction, resulting in a reduction in the amount of the characteristic gel shift. We also attempted gel shift assays using mIMCD3 nuclear extracts and the native sequence of the Tbr1 promoter (+36/+57) to see whether a relaxed cis-element for Af9 could be established. This region of the Tbr1 promoter shares sequence similarity with +78/+98 of αENaC and included in the PCR product amplified in anti-Af9 ChIP assays of Tbr1 using brain tissue (2). Despite exhaustive attempts with a variety of binding reaction conditions, we were unable to detect a sequence-specific gel shift in these assays (not shown), suggesting that the mechanisms of Af9 association with this region of the Tbr1 promoter may differ from that of αENaC, may require the chromatin context, and may not involve direct DNA binding.
Fig. 3.
Af9 binds to +78/+92 of αENaC in vitro. Gel shift (autoradiograph to the left) in which a biotinylated duplex probe was incubated with 10 μg of mIMCD3 cell nuclear extracts alone (lane 2) or in the presence of a 40-fold molar excess of unlabeled wild-type (WT) cold probe (lane 3) or mutant cold probe (Mut; lane 4). Binding reactions were also performed in the absence of nuclear extracts as a control (lane 1). Antibody interference assays (autoradiograph to the right) were performed using increasing concentrations (0.2–2 μg) of rabbit polyclonal Af9 antibody or 2 μg IgG as control. Af9 antibody decreased formation of the DNA-protein complex, whereas IgG had no effect. Gels are representative of 4 experiments each for the gel shift and for the antibody interference assays.
+78/+92 contributes to Af9-dependent, Dot1a-mediated basal suppression of αENaC promoter activity.
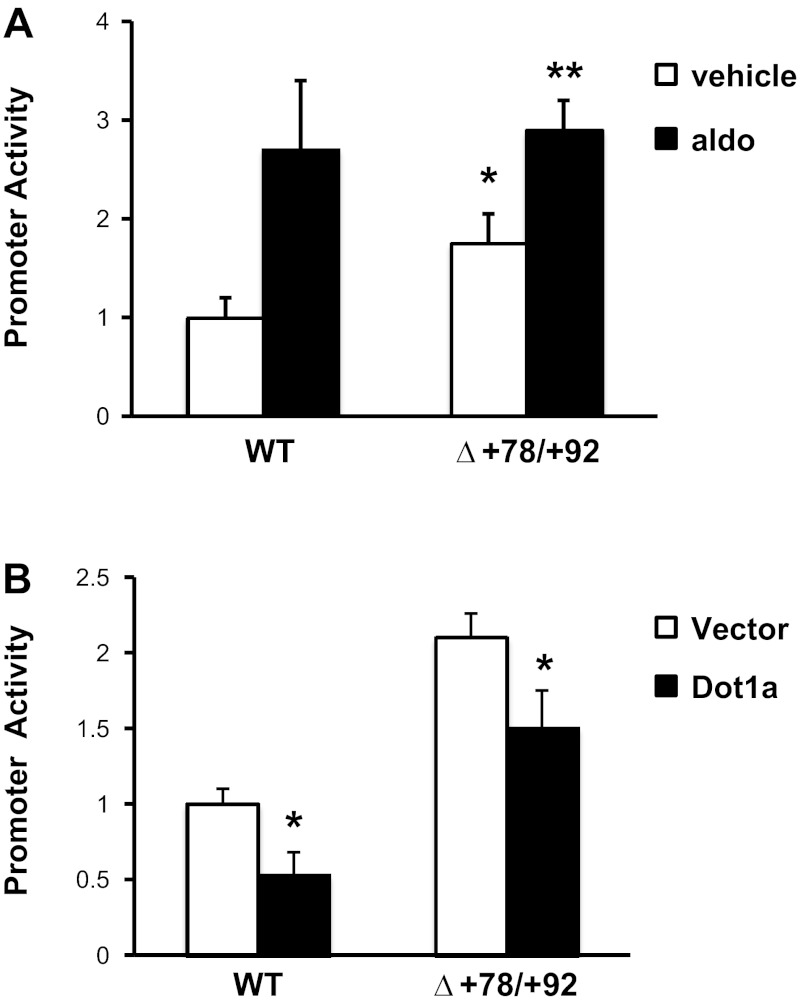
Given the key role of +78/+92 of αENaC in Af9 DNA-binding activity (Fig. 3), we next tested the functional role of +78/+92 in basal and aldosterone-induced αENaC transcription. We compared the promoter activities of the mIMCD3 cell lines stably transfected with pcDNA3.1-Zeo-1.3αENaC-Luc or pcDNA3.1-Zeo-1.3αENaCΔ+78/+92-Luc under basal conditions and after aldosterone induction. The results (Fig. 4A) revealed that the promoter activity for the cell lines harboring pcDNA3.1-Zeo-1.3αENaCΔ+78/+92-Luc was approximately twice that of the wild-type construct, approaching the levels obtained with full aldosterone induction of the wild-type promoter-reporter. This result demonstrates the functional importance of +78/+92 as an Af9-dependent negative regulatory element in αENaC promoter activity and that this element is responsible for a large proportion of the effect of aldosterone to disrupt Dot1a-Af9 repression of the αENaC promoter. Re-ChIP/qPCR failed to detect Sirt1 in anti-Af9 chromatin immunoprecipitates (data not shown), consistent with our prior finding that Sirt1 did not coimmunoprecipitate with Af9 and that Dot1a-Sirt1 and Dot1a-Af9 are separate complexes (25). Finally, since aldosterone also stimulates MR-dependent trans-activation of the αENaC promoter via the GRE at −811 (14) and disrupts Dot1a-Sirt1 repression of the αENaC promoter (25), it is not surprising that aldosterone treatment resulted in a modest, but significant further increment in pcDNA3.1-Zeo-1.3αENaCΔ+78/+92-Luc promoter activity (Fig. 4A), presumably through these alternative pathways.
Fig. 4.
+78/+92 of the αENaC promoter is critical for Dot1a-mediated basal repression and aldosterone-induced derepression of αENaC promoter activity. A: cluster of 9 point mutations was introduced into +78/+92 in pcDNA3.1-Zeo-1.3αENaC-Luc to create the pcDNA3.1-Zeo-1.3αENaCΔ+78/+92-Luc as described under materials and methods. Stable mIMCD3 cell lines harboring the pcDNA3.1-Zeo-1.3αENaC-Luc (WT) or pcDNA3.1-Zeo-1.3αENaCΔ+78/+92 (Δ+78/+92) constructs were established, cultured in DMEM/F12 plus 10% charcoal-stripped serum for 16 h, and treated with ethanol (vehicle) or 1 μM aldo for 2 h. Luciferase activities were measured and normalized to cell protein content. The value obtained for vehicle-treated cells transfected with pGL3Zeocin-1.3αENaC was set to 1, and the other conditions were normalized to it. The values shown are the mean of triplicate determinations or 4 independent experiments. Error bars indicate ± SE. *P value ≤ 0.05 vs. corresponding vehicle-treated control. B: cell lines derived in A were transiently transfected with EGFP-Dot1a or its empty expression vector. After 24 h, firefly activities were measured and normalized to cell protein content. The value obtained for vector-transfected cells harboring pcDNA3.1-Zeo-1.3αENaC-Luc (WT) was set to 1, and the other conditions were normalized to it. The values shown are the mean of triplicate determinations or 4 independent experiments. Error bars indicate ± SE. *P value < 0.05 vs. vehicle-treated WT control. **P ≤ 0.05 vs. vehicle-treated Δ+78/+92 construct.
Given our ChIP/qPCR results demonstrating impaired Dot1a association with the +74/+107-deleted promoter-reporter construct (Fig. 2B), we next sought to evaluate the functional role of the +78/+92 element in Dot1a-mediated αENaC transcriptional repression. Trans-repression experiments were performed comparing the promoter activities of the mIMCD3 cell lines stably transfected with the pcDNA3.1-Zeo-1.3αENaC-Luc or pcDNA3.1-Zeo-1Δ+78/+92-Luc constructs after transient transfection with EGFP-Dot1a or its empty vector as control. As shown in Fig. 4B, the degree of Dot1a-mediated inhibition of promoter-reporter activity was much lower in the cells expressing the +78/+92 mutations compared with the wild-type αENaC promoter (∼50 vs. ∼25%, respectively; Fig. 4B), consistent with a role of the +78/+92 element in nucleating Dot1a association and action on the αENaC promoter. Immunoblots verified comparable expression levels of the transfected EGFP-Dot1a (not shown). Again, the fact that Dot1a still suppressed pcDNA3.1-Zeo-1.3αENaCΔ+78/+92-Luc promoter activity reflects the fact that Dot1a independently interacts with Sirt1 to suppress αENaC promoter activity at other regions contained within the 1.3-kb promoter construct (25).
Time-dependent effects of aldosterone to promote Af9 and Dot1a dismissal and RNA pol II enrichment at αENaC R3.
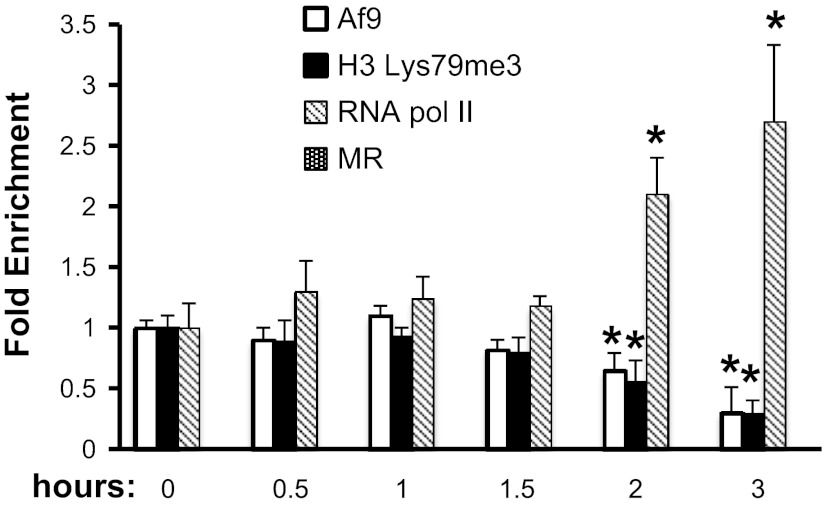
To determine the time course of aldosterone-dependent epigenetic reprogramming and transcriptional activation of αENaC, we performed a detailed ChIP/qPCR time course analysis of Af9, histone H3 trimethylated Lys79 (H3 Lys79me3, the result of Dot1a activity), RNA pol II, and MR occupancy at the αENaC R3 subregion following vehicle or aldosterone treatment of mIMCD3 cells for 0.5, 1, 1.5, 2, and 3 h. The values for the aldosterone samples were normalized to those or the time control (vehicle) samples, which did not measurably vary over the time course. The data in Fig. 5 are presented as the fold changes of each protein compared with its value at time 0. As seen in Fig. 5, significant decreases in Af9 and H3 Lys79me3 occupancy and enhanced RNA pol II occupancy at R3 were observed beginning at 2 h of aldosterone treatment. MR occupancy was not observed at any of the time points, consistent with the fact that the known GRE at −811 resides outside R3.
Fig. 5.
Time course of Dot1a-Af9 epigenetic reprogramming of the αENaC R3 subregion in mIMCD3 cells treated with aldosterone. mIMCD3 cells were cultured in DMEM/F12 plus 10% charcoal-stripped serum for 16 h and then treated with vehicle or 1 μM aldo for the indicated time periods. The cells were then harvested and subject to ChIP/qPCR assays with the antibodies specific for Af9, Dot1a, the mineralocorticoid receptor (MR), or RNA polymerase II (RNA pol II). Specific primers were used to amplify the R3 subregion of αENaC. Data from the aldo-treated samples were normalized to the vehicle time control values, which did not significantly vary over the 3 h. The data are presented as the fold change of the indicated proteins relative to their value at time 0. Error bars indicate ± SE, n = 3. *P value ≤ 0.05 vs. corresponding value at time 0.
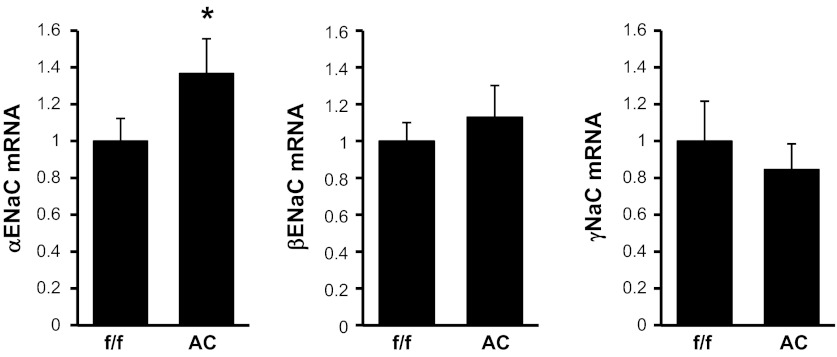
Dot1lAC mice have increased αENaC mRNA expression, despite significantly fewer principal cells.
Despite having 20% fewer principal cells in which ENaC is expressed (24), Dot1lAC mice expressed 36% greater levels of αENaC mRNA compared with Dot1lf/f mice as measured by real-time RT-qPCR (Fig. 6). The effect of CNT/CD-specific Dot1l deletion on αENaC mRNA expression in the Dot1lAC mice would likely have been even greater if the mice expressed normal numbers of principal cells. CNT/CD-specific Dot1l deletion had no measurable effect on βENaC or γENaC mRNA expression (Fig. 6). Taken together, these data support the conclusion that CNT/CD-specific genetic inactivation of Dot1l removes Dot1a-mediated repression of αENaC transcription and leads to upregulation of αENaC expression with little or no influence on βENaC and γENaC mRNA production, and they reinforce the notion that Dot1a functions as a negative regulator of basal αENaC transcription, as demonstrated in our previous work (25–28).
Fig. 6.
Connecting tubule/collecting duct-specific Dot1l inactivation in mice results in enhanced αENaC mRNA expression. mRNA levels of the indicated ENaC genes, as determined by RT-qPCR, were compared between mutant mice in which Dot1l was selectively inactivated in the connecting tubule/collecting duct (Dot1lAC) and controls from which they were derived (Dot1lf/f). f/f: Dot1lf/f. AC: Dot1lAC. Error bars indicate ± SE. *P value ≤ 0.05 vs. Dot1lf/f; n = 6 mice/genotype.
DISCUSSION
Analysis of the basal and adaptive regulation of Na+ reabsorption in the renal CD requires knowledge of transcriptional control mechanisms governing αENaC gene expression. Promoter regions of eukaryotic genes are generally composed of multiple binding sites for transcriptional activators and repressors that act in combination to regulate the expression of a linked gene. Our analysis of the αENaC gene promoter in mIMCD3 cells previously revealed that its basal activity is negatively regulated by epigenetic changes mediated by the independent interactions of Dot1a with Af9 (26–28) and with Sirt1 (25). The targeted, Af9-mediated recruitment of Dot1a to hypermethylate H3 Lys79 principally at the R3 subregion near the transcription start site of the αENaC promoter and thereby repress basal αENaC transcription appears to represent a specialized mechanism for epigenetic control, since it differs from the canonical histone H3 Lys79 methylation mark that typically localizes within the body of the gene and is associated with transcriptional activation. The present report establishes that Af9 directly binds in a sequence-specific manner to a cis-element at +78/+92 in subregion R3 of the αENaC promoter, serves as a platform for the recruitment of Dot1a to this promoter region, and thereby substantially contributes not only to the basal constraints on αENaC transcriptional activity in mIMCD3 cells, but also to the aldosterone-responsive derepression. Our gel shift/antibody competition assays indicate that Af9 binding to the +78/+92 element of αENaC is likely direct, since mutation of the +78/+92 element sequence disrupted the Af9 DNA-binding activity (Fig. 3). The binding of Af9 to this region was specific since the formation of the major complex was inhibited by the addition of excess unlabeled sequence, but not by an excess of unlabeled sequence in which the +78/+92 Af9 element was mutated (Fig. 3). The in vivo analysis, using ChIP/qPCR assays with Af9 and Dot1a antibodies and PCR primers to amplify the extended R3 fragment from mIMCD3 cells stably transfected with wild-type and +74/+107-deleted promoter-reporter constructs, confirmed the importance of +74/+107 for Af9 and Dot1a coassociation with this region and for the effect of aldosterone to disrupt the complex at this site (Fig. 2, B and C), and established that the regulatory effects of Dot1a-Sirt1 are not mediated through this Af9 element. Finally, in structure-function assays, we established that the +78/+92 site serves as a major Af9-dependent negative regulatory element for basal αENaC promoter activity (Fig. 4A), for a major part of the functional effect of Dot1a to constrain basal αENaC promoter activity (Fig. 4B), and a dominant site for aldosterone-stimulated derepression αENaC promoter activity (Fig. 4A).
Although Af9 shares sequence similarity to the yeast gene TFG3/TAF30/ANC1, a component of SWI/SNF, TFIID, and TFIIF complexes (23), relatively little is known about its specific roles in transcription. The present study, identifying for the first time Af9 binding to a gene promoter and recruitment of Dot1a, adds new mechanistic evidence. Previous structure-function analysis of the mouse αENaC gene revealed that high basal activity is retained with promoter-reporter fragments containing the 76 bp immediately downstream of the major transcription start site (12). Whether the proximity of the +78/+92 element to the transcription start site confers hindrance to the RNA polymerase II transcriptional machinery is not known. Af9 has been shown to participate in an RNA pol II transcriptional elongation complex comprised of AF4 family member proteins, Dot1a, and RNA polymerase II COOH-terminal domain kinase Cdk9/Cyclin T1/T2 (16).
Other protein-protein interactions appear to contribute to the role of Af9 in basal αENaC transcription. Evidence indicates that the interaction of Af9 with Hsp90, Hsp70, and p60/Hop is necessary for the proper subnuclear localization and activity of Af9 in mediating basal αENaC transcriptional constraint (13). However, genetic and pharmacologic manipulation of Hsp90 in those experiments had negligible effects on Af9 binding to the R3 subregion in ChIP assays (13), suggesting that this chaperone complex does not significantly contribute to the effects we observed. Af9 has been reported to interact with transcriptional corepressors, such as isoforms of the BCL6 corepressor (23) and the Polycomb homolog MPc3 (10). It is presently unknown whether Af9 also recruits these other regulatory proteins to the αENaC promoter. Finally, we also established that Af17 competes with Af9 to bind Dot1a in vitro, decreases Dot1a nuclear expression by possibly facilitating its nuclear export, and relieves Dot1a-Af9-mediated repression of αENaC promoter activity (19). Whether Af17 competes with Af9 for binding to the +78/+92 element or disrupts Dot1a-Af9 association at this site remains to be explored. However, the mechanisms regulating the Dot1a-Af9-Af17 pathway and its control of αENaC transcription may be clinically important, because mice with targeted deletion of Af17, and therefore unopposed Af9 action, exhibit renal salt wasting, hypotension, and diminished renal αENaC mRNA expression (4) as predicted from our collective results with mIMCD3 cells. Indeed, recent exploratory analyses suggest that a single nucleotide polymorphism in the human AF9 gene (MLLT3) may be associated with untreated blood pressure in African-Americans (15).
The subtle but higher levels of αENaC mRNA expression in Dot1lAC compared with Dot1lf/f mice are consistent with, but of lesser magnitude than, the values from our published cell culture data showing that siRNA-mediated knockdown of Dot1a increased αENaC mRNA levels by 57% in mIMCD3 cells (Fig. 7 in Ref. 26) and by 180% in M1 CD cells (Fig. 5 in Ref. 20). Presumably, the reduction in the principal cell population in the Dot1lAC mice might explain, in part, why Dot1lAC mice do not exhibit even greater levels of αENaC mRNA or severe impairments in salt balance (24). Moreover, the fact that the other ENaC subunit genes were not upregulated in the Dot1lAC mice (Fig. 6) suggests that the Dot1l inactivation may not substantially increase the number of functional ENaC channels. Further studies will be required to evaluate this possibility. Finally, since CNT/CD-specific inactivation of either Dot1a (this paper) or MR (21) failed to yield the significant salt-wasting phenotype that CNT/CD-specific inactivation of αENaC did (5), it will be interesting to determine whether CNT/CD-specific double knockout of MR and Dot1l reproduces the pseudohypoaldosteronism type I phenotype of CNT/CD-specific inactivation of αENaC in mice (5).
Our promoter-reporter assay results, coupled with the gel shift assay results, suggest that while +78/+92 represents the principal site of binding and trans-repressor effect of the Dot1a-Af9 complex at the αENaC promoter, other functional Af9 cis-elements may be present in the αENaC promoter. Indeed, deletion of +74/+107 did not entirely abolish Af9 and Dot1 binding to the R3 subregion in the Re-ChIP/qPCR assays (Fig. 3), and aldosterone treatment of the cells in these assays resulted in further decrements of Af9 and Dot1a binding even for the +74/+107-deleted construct (Fig. 3). However, given the minor functional contribution(s) of any such putative Af9-binding element(s) and the small contributions of other subregions to Dot1a-Af9 occupancy [<15% at R2, <5% at R1, and <5% at R0 in sequential ChIP/qPCR assays (28)] they may mediate, we have thus far been unable to discriminate any other Af9 elements from background noise.
Our time course data provide further evidence for aldosterone-induced epigenetic reprogramming that leads to a component of MR-independent derepression of αENaC transcription. ChIP/qPCR of the R3 subregion in aldosterone-treated mIMCD3 cells detected decrements in Af9 occupancy and H3 Lys79 trimethylation and increases in RNA Pol II occupancy beginning at 2 h of treatment (Fig. 5). MR occupancy of R3 was not detected over the 3-h aldosterone treatment period, consistent with evidence that the GRE resides outside this region. This time course is in agreement with our previous studies showing that mIMCD3 cells transfected with an αENaC promoter-luciferase exhibited enhanced luciferase activity by 3 h of aldosterone treatment and that endogenous αENaC mRNA levels increased within this time frame (25).
In summary, we established the importance of +78/+92 as an Af9 cis-element in directly and specifically targeting Dot1a to αENaC to mediate epigenetic control of basal and aldosterone-sensitive αENaC transcription, and as the principal mediator of the Dot1a-Af9 complex-specific effect. We also demonstrated the first in vivo evidence for the involvement of CNT/CD-expressed Dot1a in the basal repression of αENaC gene expression. Finally, our results provide the first concrete evidence for a role of Af9 as a DNA-binding protein that functions as a repressor of gene transcription. Given the roles of Af9 in cancer (22), neurodevelopmental diseases (4, 5), and potentially hypertension (4, 8), the present results may have broader implications.
GRANTS
This work was supported by National Institutes of Health Grants R01 DK075065 (to B. C. Kone), K01 DK70834 (to W. Zhang), and R01 DK080236 (to W. Zhang).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: W.Z., Z.Y., H.W., L.C., and B.C.K. conception and design of research; W.Z., Z.Y., H.W., L.C., Q.K., and B.C.K. performed experiments; W.Z., Z.Y., H.W., L.C., Q.K., and B.C.K. analyzed data; W.Z., Z.Y., H.W., L.C., Q.K., and B.C.K. interpreted results of experiments; W.Z., Z.Y., H.W., L.C., and B.C.K. prepared figures; W.Z., Z.Y., H.W., L.C., Q.K., and B.C.K. edited and revised manuscript; W.Z., Z.Y., H.W., L.C., Q.K., and B.C.K. approved final version of manuscript; B.C.K. drafted manuscript.
REFERENCES
- 1. Bhalla V, Hallows KR. Mechanisms of ENaC regulation and clinical implications. J Am Soc Nephrol 19: 1845–1854, 2008 [DOI] [PubMed] [Google Scholar]
- 2. Buttner N, Johnsen SA, Kugler S, Vogel T. Af9/Mllt3 interferes with Tbr1 expression through epigenetic modification of histone H3K79 during development of the cerebral cortex. Proc Natl Acad Sci USA 107: 7042–7047, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang MJ, Wu H, Achille NJ, Reisenauer MR, Chou CW, Zeleznik-Le NJ, Hemenway CS, Zhang W. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res 70: 10234–10242, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen L, Wu H, Pochynyuk OM, Reisenauer MR, Zhang Z, Huang L, Zaika OL, Mamenko M, Zhang W, Zhou Q, Liu M, Xia Y, Zhang W. Af17 deficiency increases sodium excretion and decreases blood pressure. J Am Soc Nephrol 22: 1076–1086, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christensen BM, Perrier R, Wang Q, Zuber AM, Maillard M, Mordasini D, Malsure S, Ronzaud C, Stehle JC, Rossier BC, Hummler E. Sodium and potassium balance depends on alphaENaC expression in connecting tubule. J Am Soc Nephrol 21: 1942–1951, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collins EC, Appert A, Ariza-McNaughton L, Pannell R, Yamada Y, Rabbitts TH. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol Cell Biol 22: 7313–7324, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corral J, Lavenir I, Impey H, Warren AJ, Forster A, Larson TA, Bell S, McKenzie AN, King G, Rabbitts TH. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell 85: 853–861, 1996 [DOI] [PubMed] [Google Scholar]
- 8. Duarte JD, Zineh I, Burkley B, Gong Y, Langaee TY, Turner ST, Chapman AB, Boerwinkle E, Gums JG, Cooper-Dehoff RM, Beitelshees AL, Bailey KR, Fillingim RB, Kone BC, Johnson JA. Effects of genetic variation in H3K79 methylation regulatory genes on clinical blood pressure and blood pressure response to hydrochlorothiazide. J Transl Med 10: 56, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haribaskar R, Putz M, Schupp B, Skouloudaki K, Bietenbeck A, Walz G, Schafer T. The planar cell polarity (PCP) protein Diversin translocates to the nucleus to interact with the transcription factor AF9. Biochem Biophys Res Commun 387: 212–217, 2009 [DOI] [PubMed] [Google Scholar]
- 10. Hemenway CS, de Erkenez AC, Gould GC. The polycomb protein MPc3 interacts with AF9, an MLL fusion partner in t(9;11)(p22;q23) acute leukemias. Oncogene 20: 3798–3805, 2001 [DOI] [PubMed] [Google Scholar]
- 11. Iida S, Seto M, Yamamoto K, Komatsu H, Tojo A, Asano S, Kamada N, Ariyoshi Y, Takahashi T, Ueda R. MLLT3 gene on 9p22 involved in t(9;11) leukemia encodes a serine/proline rich protein homologous to MLLT1 on 19p13. Oncogene 8: 3085–3092, 1993 [PubMed] [Google Scholar]
- 12. Kohler S, Pradervand S, Verdumo C, Merillat AM, Bens M, Vandewalle A, Beermann F, Hummler E. Analysis of the mouse Scnn1a promoter in cortical collecting duct cells and in transgenic mice. Biochim Biophys Acta 1519: 106–110, 2001 [DOI] [PubMed] [Google Scholar]
- 13. Lin JJ, Hemenway CS. Hsp90 directly modulates the spatial distribution of AF9/MLLT3 and affects target gene expression. J Biol Chem 285: 11966–11973, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mick VE, Itani OA, Loftus RW, Husted RF, Schmidt TJ, Thomas CP. The α-subunit of the epithelial sodium channel is an aldosterone-induced transcript in mammalian collecting ducts, and this transcriptional response is mediated via distinct cis-elements in the 5′-flanking region of the gene. Mol Endocrinol 15: 575–588, 2001 [DOI] [PubMed] [Google Scholar]
- 15. Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112: 711–723, 2003 [DOI] [PubMed] [Google Scholar]
- 16. Monroe SC, Jo SY, Sanders DS, Basrur V, Elenitoba-Johnson KS, Slany RK, Hess JL. MLL-AF9 and MLL-ENL alter the dynamic association of transcriptional regulators with genes critical for leukemia. Exp Hematol 39: 77–86, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pina C, May G, Soneji S, Hong D, Enver T. MLLT3 regulates early human erythroid and megakaryocytic cell fate. Cell Stem Cell 2: 264–273, 2008 [DOI] [PubMed] [Google Scholar]
- 18. Pramparo T, Grosso S, Messa J, Zatterale A, Bonaglia MC, Chessa L, Balestri P, Rocchi M, Zuffardi O, Giorda R. Loss-of-function mutation of the AF9/MLLT3 gene in a girl with neuromotor development delay, cerebellar ataxia, and epilepsy. Hum Genet 118: 76–81, 2005 [DOI] [PubMed] [Google Scholar]
- 19. Reisenauer MR, Anderson M, Huang L, Zhang Z, Zhou Q, Kone BC, Morris AP, Lesage GD, Dryer SE, Zhang W. AF17 competes with AF9 for binding to Dot1a to up-regulate transcription of epithelial Na+ channel alpha. J Biol Chem 284: 35659–35669, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reisenauer MR, Wang SW, Xia Y, Zhang W. Dot1a contains three nuclear localization signals and regulates the epithelial Na+ channel (ENaC) at multiple levels. Am J Physiol Renal Physiol 299: F63–F76, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ronzaud C, Loffing J, Bleich M, Gretz N, Grone HJ, Schutz G, Berger S. Impairment of sodium balance in mice deficient in renal principal cell mineralocorticoid receptor. J Am Soc Nephrol 18: 1679–1687, 2007 [DOI] [PubMed] [Google Scholar]
- 22. Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell 10: 257–268, 2006 [DOI] [PubMed] [Google Scholar]
- 23. Srinivasan RS, de Erkenez AC, Hemenway CS. The mixed lineage leukemia fusion partner AF9 binds specific isoforms of the BCL-6 corepressor. Oncogene 22: 3395–3406, 2003 [DOI] [PubMed] [Google Scholar]
- 24. Wu H, Chen L, Zhou Q, Zhang X, Berger S, Bi J, Lewis DE, Yia Y, Zhang W. Derivation of renal intercalated cells from Aqp2-expressing cells in mice. J Am Soc Nephrol In press 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang D, Li S, Cruz P, Kone BC. Sirtuin 1 functionally and physically interacts with disruptor of telomeric silencing-1 to regulate alpha-ENaC transcription in collecting duct. J Biol Chem 284: 20917–20926, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang W, Xia X, Jalal DI, Kuncewicz T, Xu W, Lesage GD, Kone BC. Aldosterone-sensitive repression of ENaCα transcription by a histone H3 lysine-79 methyltransferase. Am J Physiol Cell Physiol 290: C936–C946, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem 281: 18059–18068, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang W, Xia X, Reisenauer MR, Rieg T, Lang F, Kuhl D, Vallon V, Kone BC. Aldosterone-induced Sgk1 relieves Dot1a-Af9-mediated transcriptional repression of epithelial Na+ channel alpha. J Clin Invest 117: 773–783, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]