Abstract
STAT5a/b species are well known as transcription factors that regulate nuclear gene expression. In a novel line of research in human pulmonary arterial endothelial cells (HPAECs), we previously observed that STAT5a associated with the Golgi apparatus and that siRNA-mediated knockdown of STAT5a/b led to the rapid development of a dramatic cystic change in the endoplasmic reticulum (ER) characterized by deposition along cyst membranes and tubule-to-cyst boundaries of the proteins reticulon-4 (RTN4; also called Nogo-B) and the ER-resident GTPase atlastin-3 (ATL3) and Golgi fragmentation. We now report that STAT5a can be observed in ER sheets in digitonin-permeabilized HPAECs and that anti-STAT5a cross- immunopanned ATL3 but not RTN4. Moreover, there was marked accumulation of the 63-kDa cytoskeleton-linking membrane protein and ER-spacer CLIMP63 (also called cytoskeleton-associated protein 4, CKAP4) and KDEL-mCherry within the cysts. That the STAT5a/b-siRNA-induced cystic ER phenotype developed in the presence of the transcription inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) had suggested that the mechanism was independent of the transcription factor functions of STAT5a/b, i.e., was “nongenomic.” We have now definitively tested the requirement for the nucleus in eliciting the STAT5a/b-siRNA-induced cystic ER phenotype. Enucleated HPAEC cytoplasts were prepared using adherent 35-mm cultures using the cytochalasin B-centrifugation method (typically yielding 65–75% enucleation). STAT5a/b siRNAs readily elicited the cystic ER phenotype including the marked luminal accumulation of CLIMP63 and Golgi fragmentation in the recovered HPAEC cytoplasts demonstrably lacking a nucleus. These studies provide unequivocal evidence using enucleated cytoplasts for a nongenomic mechanism(s) underlying the cystic change in ER structure elicited by STAT5a/b knockdown.
Keywords: nongenomic functions of STAT transcription factors, pulmonary arterial endothelial cells, enucleated cytoplasts, small interfering RNA knockdown, cystic dilatation of endoplasmic reticulum, pulmonary arterial hypertension
the seven mammalian proteins comprising the “signal transducers and activators of transcription” family (STAT1, 2, 3, 4, 5a, 5b, and 6) have been investigated extensively since the mid-1990s for their ability to mediate cytokine and growth factor-induced transcriptional activation of target genes in the cell nucleus (19, 21, 30). Over the years the nuclear transcriptional regulatory function of STAT proteins, Tyr- and/or Ser-phosphorylated or even the nonphosphorylated STAT proteins, has commanded great attention (19, 21, 30). However, in recent years there is growing awareness of nongenomic functions of STAT proteins in the cytoplasm involving focal adhesions, microtubules, sequestering endosomes, and perhaps mitochondria (4, 19). In a novel line of research in human pulmonary arterial endothelial cells (HPAECs), we previously reported that STAT5a associated with the Golgi apparatus, and both STAT5a and b species were present in complexes with the endoplasmic reticulum (ER)- and Golgi apparatus-resident GTPase atlastin-3 (ATL3) (9). Dramatically, siRNA-mediated knockdown of STAT5a/b led to the rapid development within 8–12 h of a cystic change in the ER characterized by deposition of the ER proteins reticulon-4 (RTN4; also called Nogo-B) and ATL3 along cyst membranes and tubule-to-cyst boundaries, Golgi and mitochondrial fragmentation, and reduction in their respective trafficking and membrane potential functions (9). Thin-section electron microscopy (EM) confirmed the cystic dilatation of the ER with ribosomes associated with the cytosolic face of the cystic membranes as well as enlargement, dilatation, and fragmentation of the Golgi cisternae (9). The previous observation that this STAT5a/b-siRNA-induced cystic ER phenotype developed in the presence of the transcription inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB; 23) suggested that the mechanism was likely independent of the transcription factor functions of STAT5a/b, i.e., was nongenomic (9). This represented a mechanistically unexpected inference concerning a well-known transcription factor (2, 5). In light of the possibility that the inhibitory effect of DRB on mRNA synthesis might have been incomplete (23), and observations that even nonphosphorylated STAT proteins have nuclear transcriptional functions (19, 21, 30), the novelty and importance of the suggestion that nonphosphorylated STAT5 species might have nongenomic effects in maintaining the structure of the ER/Golgi organelles in the cytoplasm (9) required us to definitively test for a dependence on the cell nucleus in the development of the STAT5a/b-siRNA-induced cystic ER phenotype.
In 1972, Prescott and colleagues (17) devised a method to enucleate substrate-adherent cells using a combination of treatment with cytochalasin B and centrifugation at a tangential angle in a fixed-angle rotor. The resulting cytoplasts (still adherent to the substrate) recover rapidly and remain functional in terms of protein synthesis and nonnuclear cellular functions for the next 2–3 days (6, 11, 14–17, 27). This represents a rigorous time-tested technique for definitively evaluating the requirement for a nucleus and thus genomic functions at the single-cell level in discrete cellular mechanisms (6, 11, 14–17, 27).
In the present study, we used plastic adherent pulmonary arterial endothelial cells (HPAECs) to confirm our previous data (9) showing the association of STAT5a with the Golgi apparatus and extended these immunofluorescence studies to now show the association of STAT5a with the ER, especially with ER sheets. We discovered that the 63-kDa cytoskeleton-linking membrane protein and ER-spacer CLIMP63 (also called cytoskeleton-associated protein 4, CKAP4) accumulated markedly within STAT5a/b siRNA-induced ER cysts. Importantly, we confirmed that the development of the cystic ER phenotype by after 15–18 h (i.e., overnight) after exposure to STAT5a/b siRNA was unaffected by the RNA transcription inhibitor DRB. We then prepared enucleated HPAEC cytoplast cultures and investigated the effect of STAT5a/b siRNAs on ER structure in cells proven to be devoid of a nucleus at the single-cell level. The data obtained provide unequivocal evidence for a nongenomic basis underlying the cystic change in ER structure elicited by STAT5a/b siRNAs.
MATERIALS AND METHODS
Cell culture.
Primary HPAEC cells were purchased from Clonetics (San Diego, CA). These were seeded into T-25, T-75, six-well, or 35-mm plastic plates coated with fibronectin, collagen, and bovine serum albumin (respectively 1 μg/ml, 30 μg/ml, and 10 μg/ml in coating medium; 11). HPAECs were grown in Medium 200 supplemented with low serum growth supplement LSGS (Cascade Biologics, Carlsbad, CA) and were used between passages 4 and 10. Primary bovine PAECs (BPAECs) were used as previously described (11, 12). EA.hy926 cells, derived from primary human umbilical vein endothelial cells immortalized by fusion with A549 cells, were obtained from Dr. Michal Schwartzman (Dept. of Pharmacology, New York Medical College) and were grown in DMEM supplemented with 10% vol/vol fetal bovine serum and 100 μM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine (HAT; 11). Phase-contrast microscopy of live cultures at the conclusion of each experiment was carried out using a Nikon Diaphot Microscope and a Nikon Coolpix digital camera.
Preparation of cultures containing enucleated cytoplasts.
Adherent confluent cultures of HPAECs and BPAECs grown in coated 35-mm plastic plates were completely filled with full growth medium containing 10 μg/ml of cytochalasin B, sealed using parafilm with little to no air bubbles, and then wrapped with aluminum foil. The plates were then floated upside-down in wells of a fixed-angle Sorvall GSA rotor containing 36–48 ml of distilled water. When this was spun [11,000 rpm (19,000 g) for 50 min at 15°C] the top/lid side of each dish typically came to face the outside. Following the spin, the cultures were washed with phosphate-buffered saline, replenished with full medium, and allowed to recover at 37°C for at least 5 h prior to siRNA transfection or immunofluorescence assays. Cytoplasts were distinguished from nucleated endothelial cells by staining with 4′,6-diamidino-2-phenylindole (DAPI; 7–9).
siRNAs and plasmids.
Respective siRNA oligonucleotides were purchased from Santa Cruz Biotechnology (Santa Cruz, CA; see specific catalog numbers below). Acute knockdown of target proteins was carried out using siRNA transfections per the manufacturer's protocol. Typically, in the present experiments a total of 4 μl of a 10 μM siRNA stock and transfection reagent in a 1:1 ratio with the siRNA mixture were used per 35-mm plate or per well in a six-well plate. STAT5a/b double-knockdown experiments were carried out using 2 μl of STAT5a siRNA and 2 μl of STAT5b siRNA together with 4 μl of siRNA transfection reagent. This is an amount that produces a half-maximal cystic ER change in HPAEC cultures in terms of the number of cells affected. Corresponding controls received 4 μl of scrambled siRNA mixed with 4 μl transfection reagent. Under these conditions the scrambled siRNA transfections had no apparent effects on cells. siRNA for CLIMP63 was also purchased from Santa Cruz and used as above. The expression construct for KDEL-mCherry was a gift from Dr. Gia Voeltz (35). The expression vector for myc-tagged human CLIMP63 was purchased from Origene Technologies (Rockville, MD).
Immunofluorescence and mCherry-fluorescence microscopy of cells and cytoplasts in culture.
Cells and cytoplasts in culture were fixed using cold paraformaldehyde (4%) for 1 h and then permeabilized using a buffer containing digitonin (50 μg/ml)-sucrose (0.3 M); for comparison, the traditional Triton X-100 (0.1%) permeabilization was also used in the experiment in Fig. 1A (24). Immunofluorescence assays were carried out using antibodies from specific sources and corresponding to specific catalog numbers as enumerated below (dilution range 1:100 to 1:1,000) as described earlier (7–9). Respective immunofluorescence and mCherry fluorescence were imaged as previously reported (7, 9) using a Zeiss AxioImager M2 motorized microscopy system with Zeiss W N-Achroplan ×40/numerical aperture (NA) 0.75 or Zeiss EC Plan-Neofluor ×100/NA1.3 oil objectives equipped with an high-resolution RGB HRc AxioCam camera and AxioVision 4.8.1 software in a 1,388 × 1,040 pixel high-speed color capture mode. Controls included secondary antibodies alone, peptide competition assays, and multiple different antibodies towards the same antigen. All data within each experiment were collected at identical imaging settings.
Fig. 1.
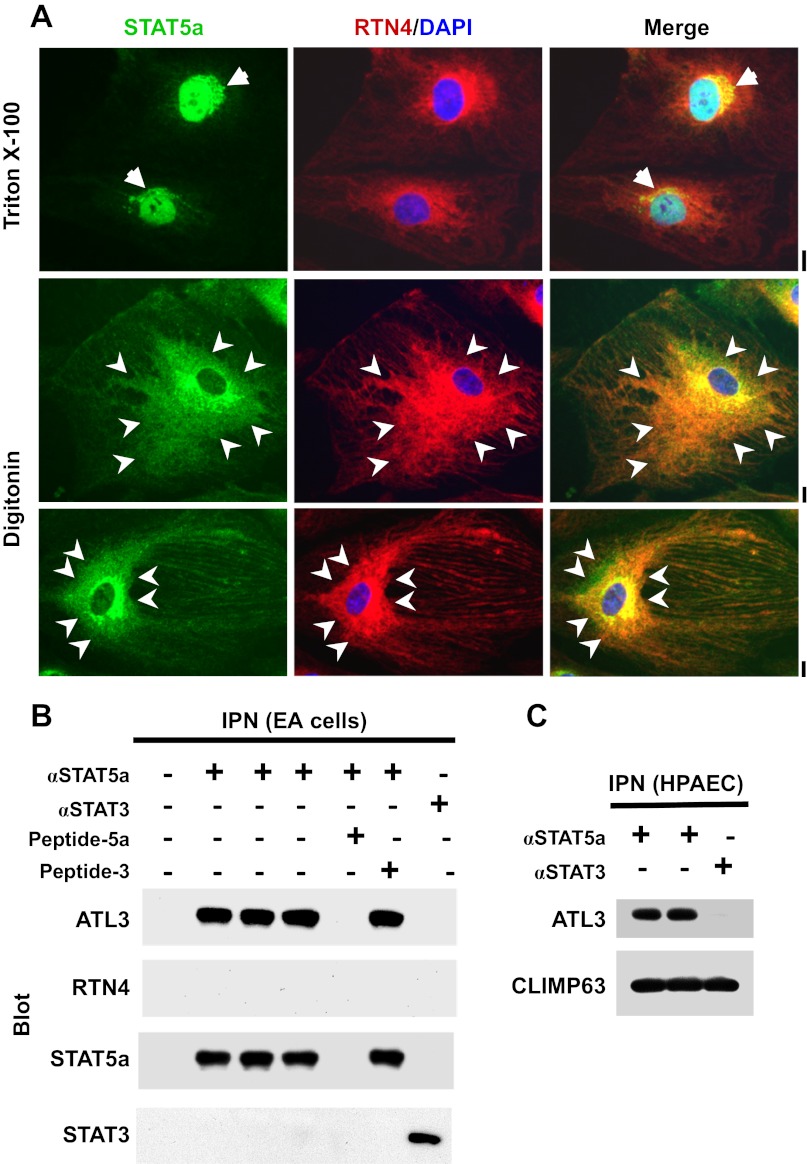
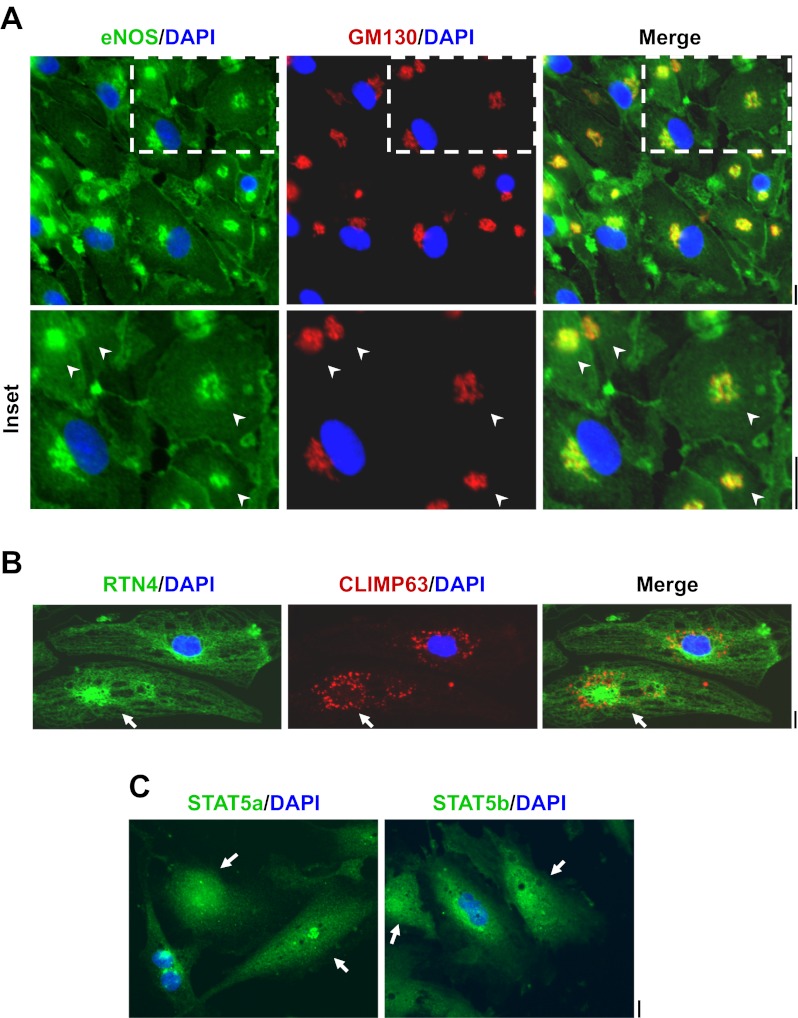
Association of signal transducer and activator of transcription 5a (STAT5a) with endoplasmic reticulum (ER) sheets and the ER-resident GTPase atlastin-3 (ATL3). A: human pulmonary arterial endothelial cell (HPAEC) cultures in 6-well plates were fixed with paraformaldehyde and then permeabilized with buffers containing either 0.1% Triton X-100 or 50 μg/ml digitonin followed by immunofluorescence imaging as indicated. Reticulon-4 (RTN4) is an ER structural protein. Arrows in the top row point to the Golgi apparatus; arrowheads in the bottom two rows highlight regions of ER sheets. Scale bars = 10 μm. B: equal aliquots of endothelial cell (EA.hy926 cells) extracts were subjected to magnetic-bead immunopanning (IPN) using anti-STAT5a pAb (in triplicate) and anti-STAT3 pAb as a negative control. The analysis included a peptide competition cross-IPN using excess of the relevant STAT5a peptide or an irrelevant STAT3 peptide. The immunoisolates were sequentially Western blotted as indicated. C: equal aliquots of HPAEC extracts were subjected to magnetic-bead IPN using anti-STAT5a or anti-STAT3 pAb. The immunoisolates were sequentially Western blotted as indicated.
Cell extracts, immunopanning, and Western blotting.
The preparation of whole cell extracts, methods for magnetic bead immunopanning using Protein A and Protein G magnetic beads (New England Biolabs, Ipswich, MA), and Western blotting were as previously reported (7–9, 22, 24, 34 and citations therein). Peptide competition experiments (as in Fig. 1B) were carried out using 5 μl of the respective pAb mixed with 20–25 μl of the stock of the competing peptide (as provided by Santa Cruz), incubation for at least 30 min at room temperature, and then using this mixture in the magnetic bead immunopanning experiment (24).
Quantitative image analyses.
This was carried out using the McMaster Biophotonics Facility version of ImageJ software (National Institutes of Health, Bethesda, MD) and respective utility plugins (available as free downloads from www.macbiophotonics.ca/imagej/) as reported previously (7–9).
Chemicals, antibodies, and siRNA reagents (respective catalog numbers in parentheses).
The mRNA synthesis inhibitor DRB was purchased from Sigma-Aldrich (D1916, St. Louis, MO) and used at 60–70 μM (26). The protein synthesis inhibitor cycloheximide (239764) was purchased from Calbiochem (Darmstadt, Germany). Cytochalasin B (sc-3519) was purchased from Santa Cruz Biotechnology and used at 10 μg/ml (6, 11, 14–17, 27). Rabbit pAbs to giantin (24586) and ATL3 (104262) was purchased from Abcam (Cambridge, MA). Rabbit pAbs to STAT3 (sc-482), STAT5a (sc-1081x), STAT5b (sc-835x), endothelial nitric oxide synthase (eNOS; sc-654), goat pAb to RTN4 (sc-11027), as well as the STAT5a peptide (sc-1081 P) and STAT3 peptide (sc-482 P) used in competition experiments were from Santa Cruz Biotechnology. Murine mAbs to GM130 (Golgi Matrix 130 kDa protein; 610823) was purchased from BD Biosciences (Eugene, OR). Anti-CLIMP63 murine mAb (804-604-C100) was purchased from Enzo Life Science (Farmingdale, NY).
Respective AlexaFluor 488- and AlexaFluor 594-tagged secondary donkey antibodies to rabbit (A-11008 and A-11012), mouse (A-21202 and A-21203), or goat (A-11055 and A-11058) IgG were from Invitrogen Molecular Probes (Eugene, OR). siRNA preparations and transfection reagents were obtained from Santa Cruz Biotechnology. These were siRNA transfection reagent (sc-29528), siRNA transfection medium (sc-36868), scrambled control-A siRNA (sc-37007), and siRNA preparations to STAT3 (h)(sc-29493), STAT5a (h)(sc-37008), STAT5b (h)(sc-370010), and CLIMP63 (h)(sc-95758).
RESULTS
Association of STAT5a with ER sheets.
When HPAEC cultures were fixed using paraformaldehyde followed by the traditional permeabilization step using Triton X-100 (0.1%), STAT5a was observed in association with the Golgi apparatus, centrosomes, and nucleus (Fig. 1A, top row; see also ref. 9). An association of STAT5a with the ER was not evident using Triton X-100 permeabilization. Moreover, when cells fixed and permeabilized using Triton were examined by thin-section EM, it became clear that this detergent was disruptive of cellular structure even in cells that had been fixed prior to permeabilization (data not shown). In contrast, when digitonin (50 μg/ml) was used for permeabilization, there was a marked preservation of cellular structure by EM (data not shown). Digitonin permeabilization resulted in increased preservation of the ER structural protein RTN4 (24, 32, 35) and its immunofluorescence (Fig. 1A, bottom two rows). This method revealed the association of STAT5a with RTN4-positive ER sheets (Fig. 1A, arrowheads), and to a lesser extent with ER tubules. However, under digitonin permeabilization conditions, anti-STAT5a pAb did not gain access to the nucleus (Fig. 1A, bottom two rows; also see ref. 24 and citations therein for detergent-dependent effects in cell preparation). Earlier we had reported that anti-ATL3 pAb, but not anti-ATL1 pAb, cross-immunopanned both STAT5a and STAT5b using extracts from EA.hy926 cells (9). The reverse of this experiment is shown in Fig. 1B in which anti-STAT5a pAb was shown to cross-immunopan ATL3. Importantly, ant-STAT5a pAb did not cross-react with RTN4 and peptide competition experiments confirmed the immunologic specificity of the interaction between STAT5a and ATL3. This interaction between anti-STAT5a pAb was also observed using extracts derived from HPAECs (Fig. 1C). Additionally, the data in Fig. 1C show that both anti-STAT5a and anti-STAT3 pAbs cross-reacted with CLIMP63 (see ref. 12 for known interactions between STAT3 and microtubule-associated proteins, especially stathmin).
Luminal content of STAT5a/b-siRNA-induced ER cysts includes CLIMP63 and KDEL-mCherry.
We have previously provided evidence for a functional role of the association of STAT5a/b species with the ER and Golgi in which HPAEC cultures were exposed to the respective siRNAs (9). When used in maximal amounts (6 μl of each siRNA per 35-mm culture), we have previously shown by Western blotting that the siRNA transfection reduces the respective STAT protein species by 80–90% by day 2 (Fig. 4C in ref. 9). By day 1, at a time when the cystic phenotype was fully developed, there was a 70–80% reduction in STAT5a/b when both siRNAs were transfected. In the present experiments we have used amounts of STAT5a and STAT5b siRNAs that give half-maximal effects (2 μl of each per 35-mm culture). In the new experiments, the STAT5a/b siRNA-induced cytoplasmic cysts (Fig. 2A) clearly represented dilated modified ER demarcated by increased deposition of the ER-resident proteins RTN4 and ATL3, the cystic membranes, and around cyst zone boundaries (Fig. 2B). However, the nature of the cyst contents remained unclear.
Fig. 4.
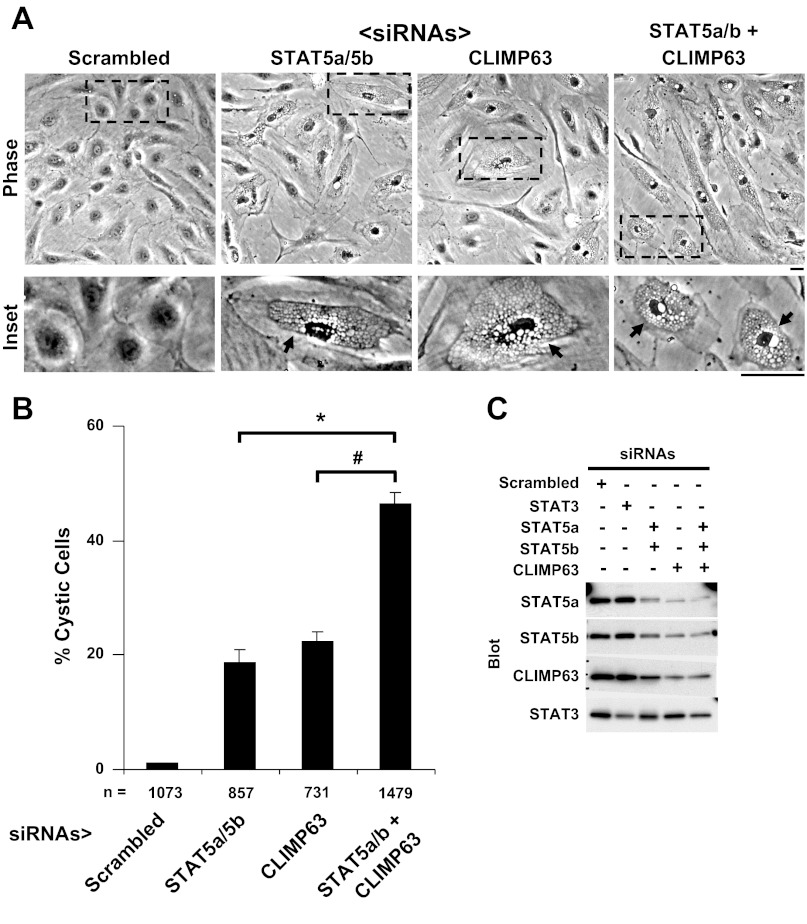
Addition of CLIMP63 siRNA enhanced development of cystic ER phenotype in response to STAT5a/b siRNAs. A: HPAEC cultures were transfected with respective siRNAs as indicated, and development of the cystic ER phenotype was evaluated 18 h later by phase microscopy. Scale bar = 25 μm. B: quantitation of % cystic cells in the respective cultures in A. n = no. of cells enumerated; error bars indicate means ± SE derived from replicate cultures (at least in triplicate). *,#P < 0.05 using Student's t-test. C: Western blot analyses showing knockdown of respective proteins by the indicated siRNAs. HPAEC cultures treated with respective siRNAs as in A for 18 h were harvested (9) and the cell extracts were evaluated for respective proteins by Western blotting of equal aliquots of total protein derived from each experimental group.
Fig. 2.
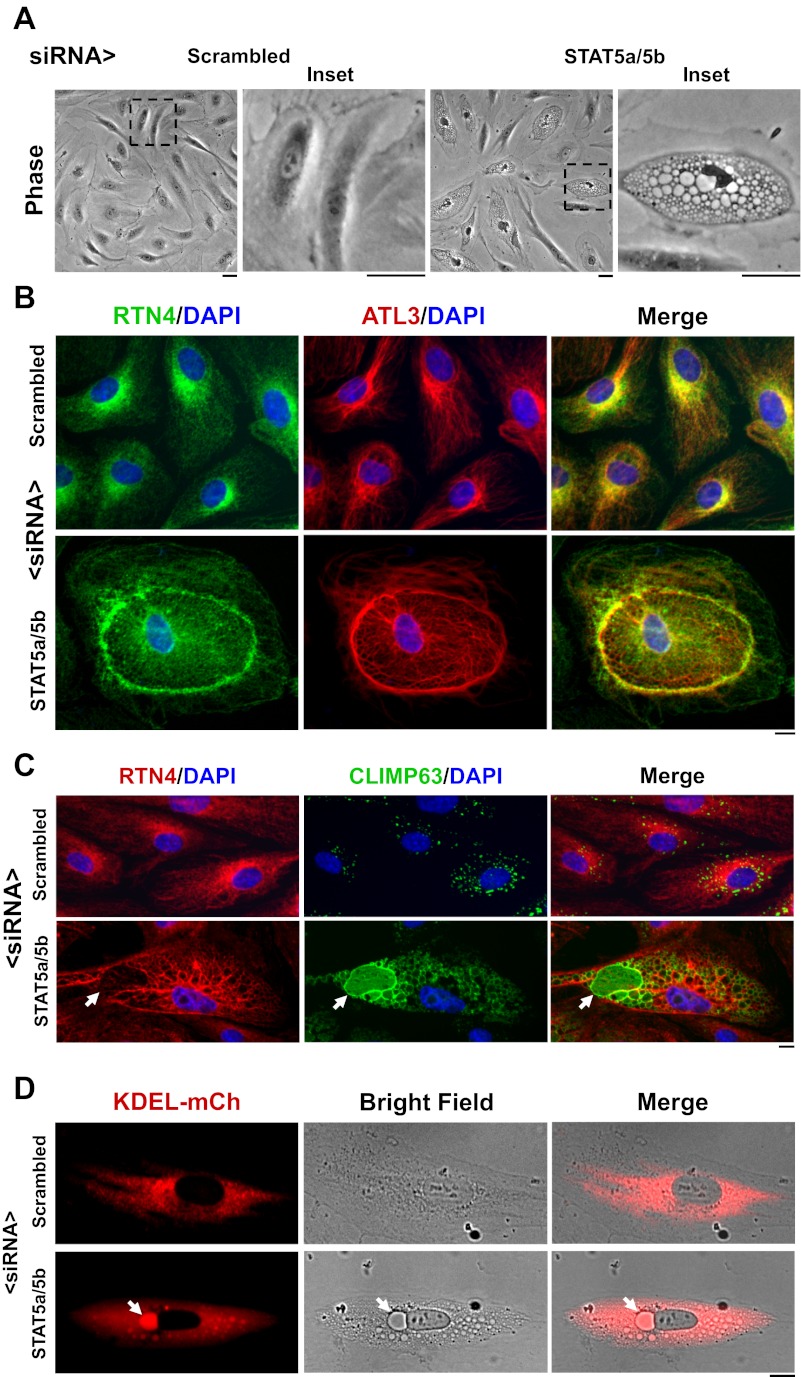
The cystic ER phenotype produced by STAT5a/b siRNAs in HPAEC cultures. A: phase-contrast images of cultures 18 h after transfection with respective siRNAs. The amount of STAT5a/b siRNAs used in this and all subsequent experiments (2 μl STAT5a siRNA + 2 μl of STAT5b siRNA per 35-mm culture and 4 μl of scrambled siRNA per 35-mm culture) was half that used for maximal cystic effect. Scale bars = 25 μm. B: immunofluorescence characterization of the cystic ER phenotype showing the cystic change and increased deposition of RTN4 and ATL3 at the cyst-zone boundaries. Scale bar = 10 μm. C: accumulation of CLIMP63 in cyst membranes and in the intracystic lumen (arrows). Scale bar = 10 μm. D: live-cell images showing increased accumulation of KDEL-mCherry (mCh) in the intracystic lumen (arrows). Scale bar = 10 μm.
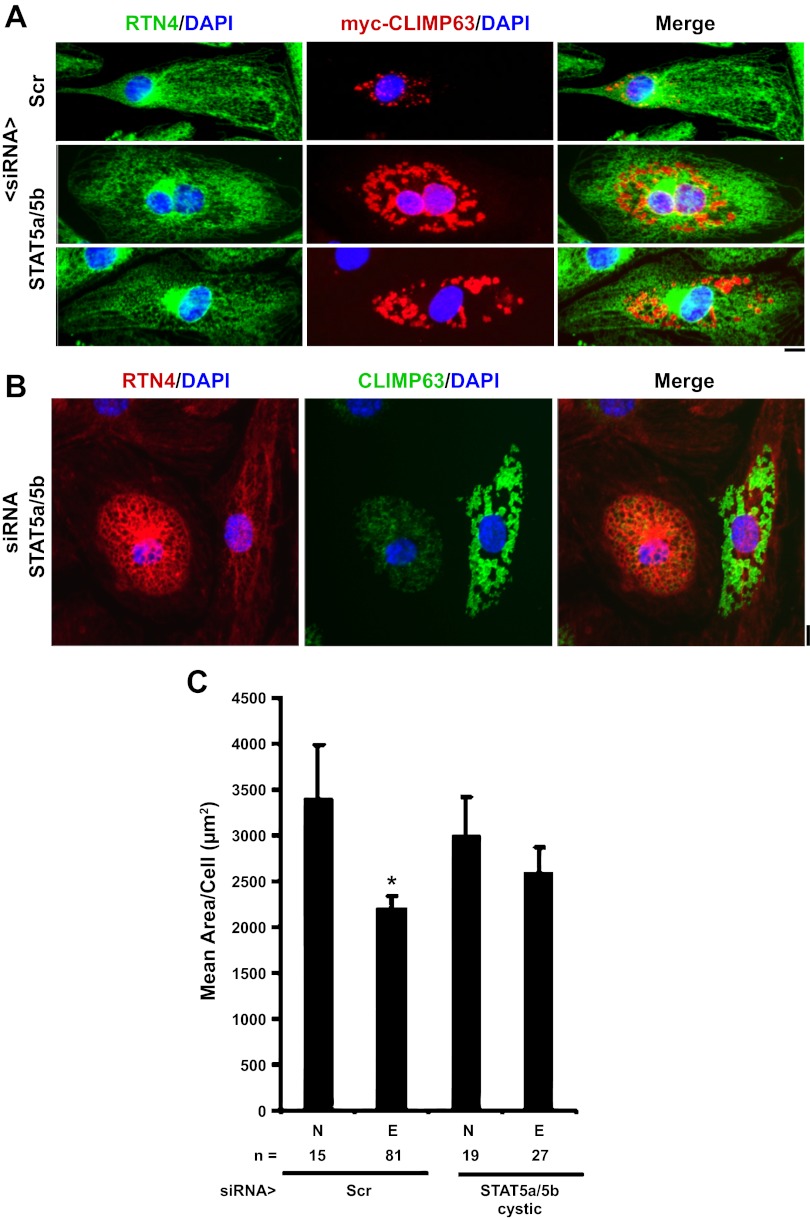
Figure 2C provides evidence showing the intracystic luminal accumulation of the “ER-spacer” and ER-to-microtubule tether protein CLIMP63 as well as along intercystic membranes (Fig. 2C; 10, 13, 25). Figure 2C shows the reorganization of endogenous CLIMP63; exogenously expressed myc-tagged CLIMP63 also accumulated in the intracystic space (Fig. 3A). Figure 2D shows live-cell images evidencing the accumulation of the ER luminal marker KDEL-mCherry (35) also in the intracystic lumen. These data demonstrate that the STAT5a/b siRNA-induced cysts represented dilatation of the ER lumen with preservation of its vectorial protein synthesis and accumulation properties. However, there was a range of variation at the single-cell level in the relative protein changes in cystic cells (Fig. 3, A and B). While cystic cells clearly showed endogenous CLIMP63 and exogenous myc-CLIMP63 within cystic lumens, not all cells showed the increased association of this protein with cystic membranes (compare Figs. 2C, 3A, and 3B). The overall size of noncystic or cystic cells in primary HPAEC cultures was not significantly different in cultures transfected with scrambled or STAT5a/b siRNAs (Fig. 3C and data not shown).
Fig. 3.
Variations in the cystic phenotype produced by STAT5a/b siRNAs in nucleated HPAECs. A: HPAECs were transfected with the respective siRNAs as indicated together with an expression vector for myc-CLIMP63. Cellular phenotype was evaluated using an antibody for endogenous RTN4 and an anti-myc mAb to label the exogenously expressed myc-CLIMP63. Scr, scrambled. Scale bar = 10 μm. B: variation in endogenous RTN4 and CLIMP63 immunofluorescence in two cystic cells side-by-side in the same culture transfected with STAT5a/b siRNAs. Scale bar = 10 μm. C: variations in cell size of nucleated or enucleated cells with or without the cystic phenotype as measured using phase-contrast microscopy and ImageJ (7, 8). n = no. of cells sized in each group; N, nucleated cells; E, enucleated cells. *P < 0.05.
Additionally, Fig. 4A shows that CLIMP63 siRNA by itself led to development of a cystic ER phenotype, and Fig. 4B shows that the combination of CLIMP63 and STAT5a/b siRNAs (each used at the half-maximal 2 μl amount per 35 mm culture) was additive, and produced a cystic ER phenotype in up to 40–50% of the cells in a HPAEC culture. Importantly, Fig. 4C confirms that the respective STAT5a/b and CLIMP63 siRNAs knocked down the respective proteins in HPAECs. As a negative control, STAT3 siRNA did not affect STAT5a/b or CLIMP63 levels. However, siRNA to CLIMP63 did reduce levels not only of CLIMP63 but also of STAT5a and b, consistent with the observation of CLIMP63 in immunoisolatable complexes with STAT5a (Fig. 1C). Overall, the available evidence (Figs. 1–4) shows that STAT5 species interface with the ER-resident proteins ATL3 and CLIMP63. While these data set the stage for exploration of the detailed interactions between STAT5 species and ATL3 and CLIMP63 at the level of the ER membranes, nevertheless, the critical mechanistic question was whether the development of the cystic ER phenotype resulting from acute STAT5a/b knockdown in HPAECs involves any transcriptional/nuclear functions of STAT5.
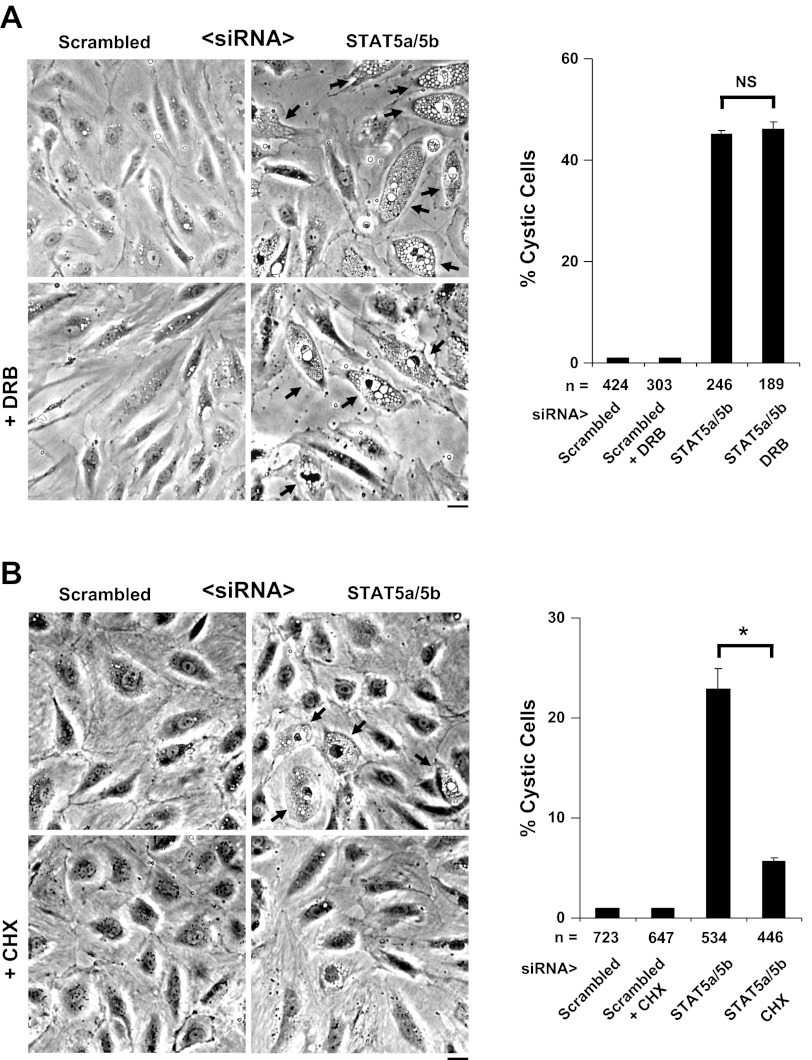
Development of the STAT5a/b siRNA-induced cystic ER phenotype is unaffected by DRB but is reduced by cycloheximide.
Exposure of STAT5a/b siRNA-treated HPAEC cultures to the mRNA synthesis inhibitor DRB did not affect the development of the cystic ER phenotype (Fig. 5A), confirming our prior observation (Fig. 7B in ref. 9). On the basis of data such as these we had suggested that the effect of STAT5a/b siRNA in eliciting a cystic ER phenotype was nongenomic, i.e., did not involve a transcriptional or nuclear mechanism (9). In contrast, the data in Fig. 5B show that the protein synthesis inhibitor cycloheximide reduced development of the cystic phenotype consistent with the need for ongoing vectorial protein synthesis for resulting in the observed ER dilatation (see Figs. 2C, 2D, 3A, and 3B above for vectorial accumulation of lumen-resident proteins within ER cysts). Since inhibitors such as DRB could be leaky or incomplete (23), the critical issue was to obtain unequivocal evidence for the involvement of the nucleus or not in the development of the STAT5a/b siRNA-induced cystic ER phenotype.
Fig. 5.
Development of cystic ER phenotype is unaffected by 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) but is inhibited by cycloheximide (CHX). A: HPAEC cultures were transfected with the respective siRNAs in the presence of DRB (75 μM) beginning 5 h before the transfection and the inhibitor was maintained in the cultures overnight. Development of the cystic ER phenotype was assessed by phase-contrast microscopy and quantitated as in Fig. 4B (means ± SE; NS = not significant; P > 0.05 using Student's t-test). Arrows highlight cystic cells. Scale bar = 25 μm. B: HPAEC cultures were transfected with the respective siRNAs in the presence of CHX (5 μg/ml) beginning 5 h before the transfection and the inhibitor was maintained in the cultures overnight. Development of the cystic ER phenotype was assessed by phase-contrast microscopy and quantitated as in Fig. 4B (means ± SE; *P < 0.05 using Student's t-test). Arrows highlight cystic cells. Scale bar = 25 μm.
Fig. 7.
Development of the cystic ER phenotype is unimpaired in enucleated HPAEC cytoplasts upon exposure to STAT5a/b siRNAs. Adherent HPAECs grown in 35-mm plates were enucleated using the cytochalasin B-centrifugation method, allowed to recover for 5 h, and then transfected with the respective siRNAs. After overnight incubation, triple-color imaging was carried out for the ER structural protein RTN4, the intracystic ER protein CLIMP63 (see Fig. 2C), giantin (Golgi marker), and DAPI as a nuclear marker as indicated. A: typical data at the single-cell level, with enucleated cells indicated by arrowheads (scale bar = 10 μm). B: the respective quantitation derived from 3 independent experiments (mean ± SE; NS = not significant; P > 0.05 using Student's t-test). C: a compact Golgi in a cytoplast exposed to scrambled siRNA (arrow in top row), as well as a fragmented Golgi in a cytoplast exposed to STAT5a/b siRNAs (arrow in bottom row). Scale bar = 10 μm.
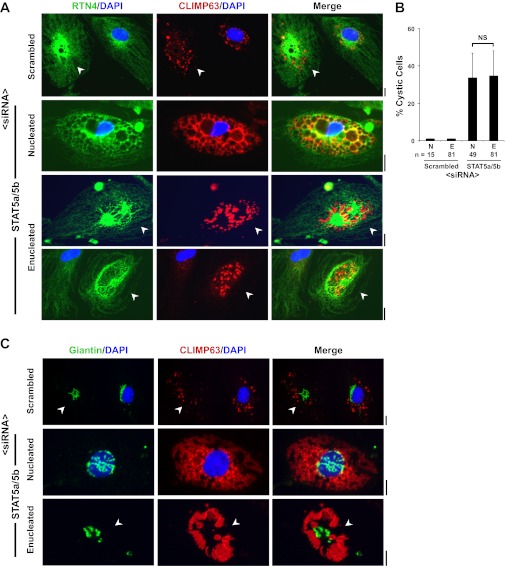
STAT5a/b siRNAs elicit a cystic ER phenotype in enucleated HPAEC cytoplasts.
The cytochalasin B-centrifugation method of Prescott and colleagues (17) was optimized for the preparation of enucleated HPAEC cytoplasts in adherent cultures grown in 35-mm coated plastic petri dishes. Typically, 65–75% of the adherent cells were enucleated as judged by subsequent DAPI staining. After an overnight recovery period such nucleus-free cytoplasts prepared from HPAECs contained a compact Golgi (Fig. 6A), maintained localization of eNOS in both the plasma membrane and Golgi apparatus (Fig. 6A), maintained the normal structure of the ER as evidenced by RTN4 and CLIMP63 immunofluorescence (Fig. 6B) and persistent expression of STAT5a and STAT5b (Fig. 6C). With respect to control HPAECs, there was a significant reduction in cell size upon enucleation (Fig. 3C).
Fig. 6.
Characterization of enucleated endothelial cell cytoplasts. A–C: adherent HPAEC cultures in 35-mm plates were subjected to the cytochalasin B-centrifugation enucleation procedure (typical yield 65–75% enucleation), allowed to recover overnight, and then immunostained for GM130 (Golgi marker), RTN4 (ER marker), endothelial nitric oxide synthase (eNOS), CLIMP63, STAT5a, and STAT5b and then stained for DAPI (nucleus marker) as indicated. Illustrative data are shown. Arrowheads in A and arrows in B and C point to nucleus-free cytoplasts. Scale bars = 25 μm in A and 10 μm in B and C.
For siRNA exposure, HPAECs in 35-mm dishes were subjected to cytoplast preparation, allowed to recover for 5 h, and then exposed to STAT5a/b siRNAs or the control scrambled siRNA preparations. The cytoplasts were then incubated overnight (15–18 h) in full HPAEC growth medium. Figure 7, A and B, shows that STAT5a/b siRNAs elicited a cystic ER phenotype equally well in both nucleated and nucleus-free cytoplasts. It is unequivocal that nucleus-free cytoplasts respond to STAT5a/b siRNAs with development of the cystic ER phenotype (Fig. 7A, arrowheads). Figure 7B represents the pooled numerical data from three independent experiments evaluated using phase-contrast microscopy of cystic cells; there is no difference in the cystic ER response to STAT5a/b siRNAs between nucleated cells or cytoplasts. Figure 7C extends these observations to the determination that nucleus-free cytoplasts which show a cystic ER (accumulation of CLIMP63 in the ER cysts) also show Golgi fragmentation. Finally, the data in Fig. 8 illustrate the range of variation that can be observed in CLIMP63 accumulation in cystic enucleated cultures of heterogeneous HPAECs; in some cystic enucleated cells the CLIMP63 appears only in cyst lumens (Fig. 8, middle row), in others in both the lumen and the cystic membranes (Fig. 8, bottom row).
Fig. 8.
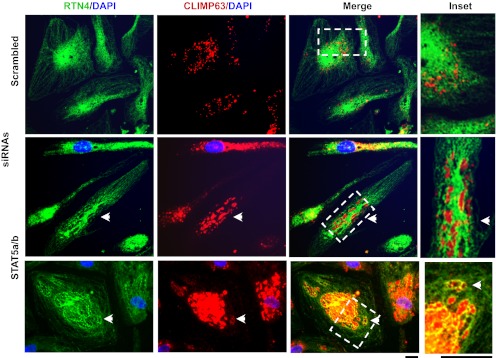
Variations in the characteristics of the cystic phenotype produced by STAT5a/b siRNAs in enucleated HPAECs. Cultures of enucleated HPAECs in 35-mm plates were transfected with scrambled or STAT5a/b siRNAs and characterized 1 day later by immunofluorescence as indicated. The cells illustrated (arrowheads) show the extent of variation in CLIMP63 distribution in cystic cells (within cyst lumen only in the middle row and within cyst lumen plus in cystic membranes in the bottom row). Scale bars, 10 μm.
DISCUSSION
Following the discovery of the association of STAT5a with the Golgi apparatus and that acute siRNA-mediated knockdown of the well-known transcription factors STAT5a/b led to an extensive cystic change in ER structure and to Golgi fragmentation (9), the question of whether manifestations of this phenotype were dependent on alterations in the transcriptional/nuclear functions of STAT5 species became a critical mechanistic issue which required an unequivocal answer. The previous data showing that STAT5a and STAT5b were present in complexes which included the ER-resident GTPase ATL3 and that development of the cystic ER phenotype was unaffected by the transcriptional inhibitor DRB suggested a nongenomic basis for the development of the cystic ER/fragmented Golgi phenotype in response to STAT5a/b siRNAs (9). The present study definitively confirms this previous suggestion. Enucleated HPAEC cytoplasts, provably lacking a DAPI-stainable nucleus at the single-cell level, responded to STAT5a/b siRNAs to rapidly yield a cystic ER/fragmented Golgi phenotype. Thus, whatever the underlying mechanisms that link the acute knockdown of STAT5a/b proteins to these changes in organellar structure, such mechanisms must be nongenomic. Moreover the new data implicate the luminal accumulation of CLIMP63 in mechanisms that lead to the development of the STAT5a/b-siRNA induced ER cysts.
From a broader perspective, in recent years there has been increasing discussion of possible nongenomic functions of STAT protein family members in the cytoplasm (4, 19, 30). This is not surprising in that STAT family members largely have a cytoplasmic provenance, with only a small fraction cycling through the nucleus (perhaps up to 15% of each member; ref. 19). Examples of cytokine-activated targeting of Tyr-phosphorylated STAT3 (PY-STAT3) to cytoplasmic structures for likely functions in the cytoplasm are represented by the targeting of PY-STAT3 to focal adhesions (26) and to cytoplasmic sequestering endosomes (34). Moreover, nonphosphorylated STAT3 has been implicated to affect microtubule function in the cytoplasm by associating with the microtubule-regulatory protein stathmin (12). More recently, it has been suggested that STAT3 associated with mitochondria and thus affects mitochondrial function in a nongenomic manner (3, 33). However, the association of STAT3 with mitochondria remains controversial (1), and it is not clear whether the effects of STAT3 on mitochondrial function are unequivocally nongenomic. In contrast, the present data obtained using enucleated HPAEC cytoplasts unequivocally demonstrate that the effects on ER and Golgi organellar structure of an acute knockdown of STAT5a/b do not require the cell nucleus.
The present data show that the traditional immunofluorescence method that uses buffers containing Triton X-100 to permeabilize fixed cells leaches out considerable cytoplasmic protein and fails to reveal the association of STAT5a with ER sheets (Fig. 1A, top row). The alternative approach of using a low-concentration digitonin buffer allowed the association of STAT5a with ER sheets to be visualized (Fig. 1A, bottom two rows). The association of STAT5a with the Golgi apparatus was evident using either method (Fig. 1A, all rows) while the constitutive nuclear presence of STAT5a was evident only after Triton permeabilization (Fig. 1A, top row). Thus, the present studies, which use digitonin-permeabilization, provide immunofluorescence evidence for the association of STAT5a with the ER (Fig. 1A, bottom two rows). Moreover, the magnetic-bead immunopanning experiments further confirmed the specific association of STAT5a in complexes with the ER-resident GTPase ATL3 (Fig. 1B). These data lead to the question of how STAT5 species might constitutively affect ATL3 function and thus ER structure.
The acute knockdown of STAT5a/b using respective siRNAs led to a rapid and dramatic cystic change in ER structure (Fig. 2 and ref. 9). The question of whether the fluid compartment within these cysts contained proteins that are ordinarily resident within the ER lumen was answered by the observations that CLIMP63 and KDEL-mCherry accumulated in the intracystic space (Figs. 2C, 2D, 3A, and 3B). Moreover, increased levels of CLIMP63 were also associated with the cyst membranes in at least some cells (Fig. 2C). Two separate functions have been previously attributed to CLIMP63: 1) as an ER-to-microtubule tether on the cytosolic face of the ER (13) and 2) as an ER-spacer on the luminal side such that two CLIMP63 molecules associated with adjacent ER membranes homodimerize in trans across the space of the ER lumen locking the two adjacent membranes a fixed distance apart (45–50 nm; 10, 25). The accumulation of CLIMP63 in intracystic spaces in STAT5a/b siRNA-treated HPAECs suggests disruption of the ER-spacer function of this protein. A functional relationship between CLIMP63 biology and that of STAT5a/b at the ER became evident from the observation that CLIMP63 siRNA also produced a cystic ER phenotype (Fig. 4). Moreover, the combination of STAT5a/b and CLIMP63 siRNAs was even more effective in producing a cystic phenotype than either one alone (Fig. 4). This effect was selective in that siRNAs to ATL3, RTN4, or STAT3 neither produced the cystic phenotype nor affected that produced by STAT5a/b siRNAs (9), and CLIMP63 was observed in complexes with STAT5a (Fig. 1C). Additionally, that both CLIMP63 and the ER luminal marker KDEL-mCherry accumulated in intracystic spaces indicated that ER-associated protein synthesis and vectorial transport of the synthesized proteins to the ER lumen continued to take place in the ER cysts. That the protein synthesis inhibitor cycloheximide reduced development of the cystic ER phenotype (Fig. 5B) suggests the mechanistic dependence on vectorial protein accumulation in the ER lumen to culminate in the development of the cystic ER phenotype.
The cytochalasin B-centrifugation method (17) was readily applicable to adherent cultures of primary HPAECs grown in 35-mm dishes. Enucleated cytoplasts showed a normal ER structure and a compact Golgi apparatus (Fig. 6). It was thus possible 5 h later to expose such cultures to respective siRNAs and to monitor development of organellar changes at the single-cell level in DAPI-negative cytoplasts. The data show that development of the cystic ER phenotype in response to acute STAT5a/b siRNA-knockdown (Fig. 7) was not dependent on the presence of a nucleus. Thus STAT5a/b modulated ER structure independent of nuclear transcription.
The STAT5-dependent cytoplasmic organelle-related mechanisms are relevant to discussions of the pathogenesis of idiopathic pulmonary arterial hypertension (IPAH), especially why this disease is two to four times more prevalent in women (the so-called gender bias; 18, 31). Over 30 years ago, Smith and Heath reported observing cystic dilatation of the ER in pulmonary arterial endothelial cells in disease lesions in human IPAH and in the hypoxia model of PAH in the rat (28, 29). Our data showing development of the STAT5a/b siRNA-induced cystic ER phenotype in HPAECs replicated these observations in endothelial cells in culture (9). Importantly, STAT5 species are well known to bind to the estrogen receptor and be modulated by estradiol-17β (5, 21). Thus, our new data lead to the question of whether estradiol also modulates the novel nongenomic functions of STAT5 species at the level of ER/Golgi structure and trafficking functions, perhaps accounting for the increased prevalence of IPAH in women (20).
To summarize, the present study using enucleated HPAEC cytoplasts provides definitive evidence showing that STAT5 species regulate ER structure in the cytoplasm through nonnuclear mechanisms.
GRANTS
This work was supported by National Institutes of Health Grants R01 HL-087176 (to P. B. Sehgal) and F31 HL-107013 (to J. E. Lee).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
J.E.L., H.Y., and P.B.S. conception and design of research; J.E.L., Y.-M.Y., and H.Y. performed experiments; J.E.L., Y.-M.Y., and P.B.S. analyzed data; J.E.L., Y.-M.Y., and P.B.S. interpreted results of experiments; J.E.L. and P.B.S. prepared figures; J.E.L. and P.B.S. drafted manuscript; J.E.L. and P.B.S. edited and revised manuscript; J.E.L. and P.B.S. approved final version of manuscript.
REFERENCES
- 1. Cimica V, Chan HC, Iyer JK, Reich NC. Dynamics of the STAT3 transcription factor: nuclear import dependent on Ran and importin-β1. PLos One 6: e20188, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J 13: 4361–4369, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324: 1713–1716, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gough DJ, Sehgal P, Levy DE. Nongenomic functions of STAT3. In: JAK-STAT Signaling: From Basics to Disease, edited by Decker T, Muller M. New York: Springer-Verlag, 2012 [Google Scholar]
- 5. Hennighausen L, Robinson GW. Interpretation of cytokines signaling through the transcription factors STAT5A and STAT5B. Genes Dev 22: 711–721, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaufman Y, Berke G. Enucleated cytotoxic T lymphocytes bind specifically to target cells in vitro. Transplantation 29: 374–378, 1980 [DOI] [PubMed] [Google Scholar]
- 7. Lee JE, Patel K, Almodovar S, Tuder RM, Flores SC, Sehgal PB. Dependence of Golgi apparatus integrity on nitric oxide in vascular cells: implications in pulmonary arterial hypertension. Am J Physiol Heart Circ Physiol 300: H1141–H1158, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee JE, Reich R, Xu F, Sehgal PB. Golgi, trafficking, and mitosis dysfunctions in pulmonary arterial endothelial cells exposed to monocrotaline pyrrole and NO scavenging. Am J Physiol Lung Cell Mol Physiol 297: L715–L728, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JE, Yang YM, Liang FX, Gough DJ, Levy DE, Sehgal PB. Nongenomic STAT5-dependent effects on Golgi apparatus and endoplasmic reticulum structure and function. Am J Physiol Cell Physiol 302: C804–C820, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin S, Sun S, Hu J. Molecular basis for sculpting the endoplasmic membrane. Int J Biochem Cell Biol 44: 1436–1443, 2012 [DOI] [PubMed] [Google Scholar]
- 11. Mandic A, Hanson J, Linder S, Shoshan MC. Cisplatin induces endoplasmic reticulum stress and nucleus-independent apoptotic signaling. J Biol Chem 278: 9100–9106, 2008 [DOI] [PubMed] [Google Scholar]
- 12. Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, Cao X. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol 172: 245–257, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nikonov AV, Hauri HP, Lauring B, Kreibisch G. Climp-63-mediated binding of microtubules to the ER affects the lateral mobility of translocon complexes. J Cell Sci 120: 2248–2258, 2007 [DOI] [PubMed] [Google Scholar]
- 14. Panaretakis T, Hjortsberg L, Tamm KP, Bjorklund AC, Joseph B, Grander D. Interferon α induces nucleus-independent apoptosis by activating extracellular signal-regulated kinase 1/2 and c-Jun NH2-terminal kinase downstream of phosphatidylinositol 3-kinase and mammalian target of rapamycin. Mol Biol Cell 19: 41–50, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poste G. Enucleation of mammalian cells by cytochalasin B. I Characterization of enucleate cells. Exp Cell Res 73: 273–286, 1972 [DOI] [PubMed] [Google Scholar]
- 16. Poste G, Reeve P. Enucleation of mammalian cells by cytochalasin B. II. Fusion of hybrid cells and heterokaryons by fusion of anucleate and nucleated cells. Exp Cell Res 73: 287–294, 1972 [DOI] [PubMed] [Google Scholar]
- 17. Prescott DM, Myerson D, Wallace J. Enucleation of mammalian cells with cytochalasin B. Exp Cell Res 71: 480–485, 1972 [DOI] [PubMed] [Google Scholar]
- 18. Runo JR, Loyd JE. Primary pulmonary hypertension. Lancet 361: 1533–1544, 2003 [DOI] [PubMed] [Google Scholar]
- 19. Sehgal PB. Paradigm shifts in STAT signaling. Semin Cell Dev Biol 19: 329–340, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sehgal PB, Lee JE. Protein trafficking dysfunctions: role in the pathogenesis of pulmonary arterial hypertension. Pulm Circ 1: 17–32, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sehgal PB, Levy DE, Hirano T. Signal Transducers and Activators of Transcription (STATs): Activation and Biology. Boston, MA: Kluwer Acad., 746 pp, 2003 [Google Scholar]
- 22. Sehgal PB, Mukhopadhyay S, Xu F, Patel K, Shah M. Dysfunctions of Golgi tethers, SNAREs, and SNAPs in monocrotaline-induced pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol 292: L1526–L1542, 2007 [DOI] [PubMed] [Google Scholar]
- 23. Sehgal PB, Tamm I, Darnell JE., Jr The inhibition by DRB (5,6-dichloro-1-β-d-ribofuranasylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell 9: 473–480, 1976 [DOI] [PubMed] [Google Scholar]
- 24. Shah M, Patel K, Mukhopadhyay S, Xu F, Guo G, Sehgal PB. Membrane-associated STAT3 and Py-STAT3 in the cytoplasm. J Biol Chem 281: 7302–7308, 2006 [DOI] [PubMed] [Google Scholar]
- 25. Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell 143: 774–788, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res 64: 3550–3558, 2004 [DOI] [PubMed] [Google Scholar]
- 27. Siroky J. Comparison of the coverslip and the discontinuous percoll density gradient methods of enucleation of mouse cells. Gen Physiol Biophys 3: 119–126, 1984 [PubMed] [Google Scholar]
- 28. Smith P, Heath D. Ultrastructure of hypoxic hypertensive pulmonary vascular disease. J Pathol 121: 93–100, 1977 [DOI] [PubMed] [Google Scholar]
- 29. Smith P, Heath D. Electron microscopy of the plexiform lesion. Thorax 34: 177–186, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stark GR, Darnell JE. The JAK-STAT pathway at twenty. Immunity 36: 503–514, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tuder RM, Marecki JC, Richter A, Fijalkowska I, Flores S. Pathology of pulmonary hypertension. Clin Chest Med 28: 23–42, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586, 2006 [DOI] [PubMed] [Google Scholar]
- 33. Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang O, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu XY, Avadhani N, Drake JI, Fawcett P, Lesnefsky EF, Larner AC. Function of mitochondrial Stat3 in cellular respiration. Science 17: 475–479, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu F, Mukhopadhyay S, Sehgal PB. Live cell imaging of interleukin-6 induced targeting of “transcription factor” STAT3 to sequestering endosomes in the cytoplasm. Am J Physiol Cell Physiol 293: C1374–C1382, 2007 [DOI] [PubMed] [Google Scholar]
- 35. Zurek N, Sparks L, Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic 12: 28–41, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]