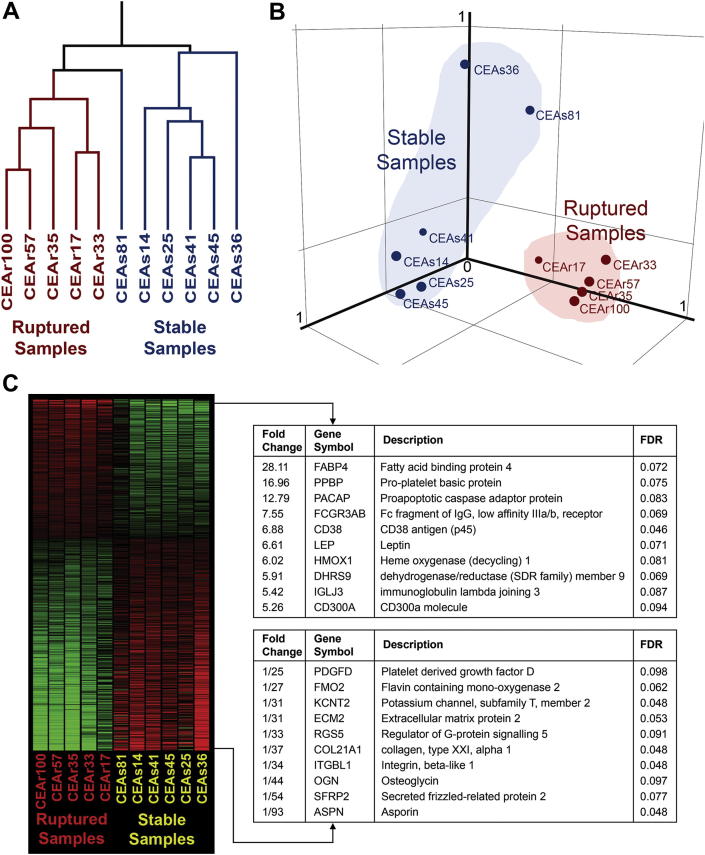
Fig. 2.
Unsupervised explorative clustering on the entire array datasets of the samples using condition tree (Panel A) and principal components analysis (Panel B). The ruptured group cluster closely together; the stable group, although clearly separate from the ruptured group, has a greater spread (as might be expected in a histologically more heterogeneous group). In panel C, statistically significant differentially expressed genes (1187 probesets representing 914 different genes) ranked by fold difference and represented in a heatmap is shown. Red represents over-expression, green represents under-expression. The top 10 differentially expressed genes from both ends are listed. Fold change values above 1 represents higher relative expression in the ruptured group and values below 1 represents higher relative expression in the stable group. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)