Figure 2.
Summary of Analysis Methods
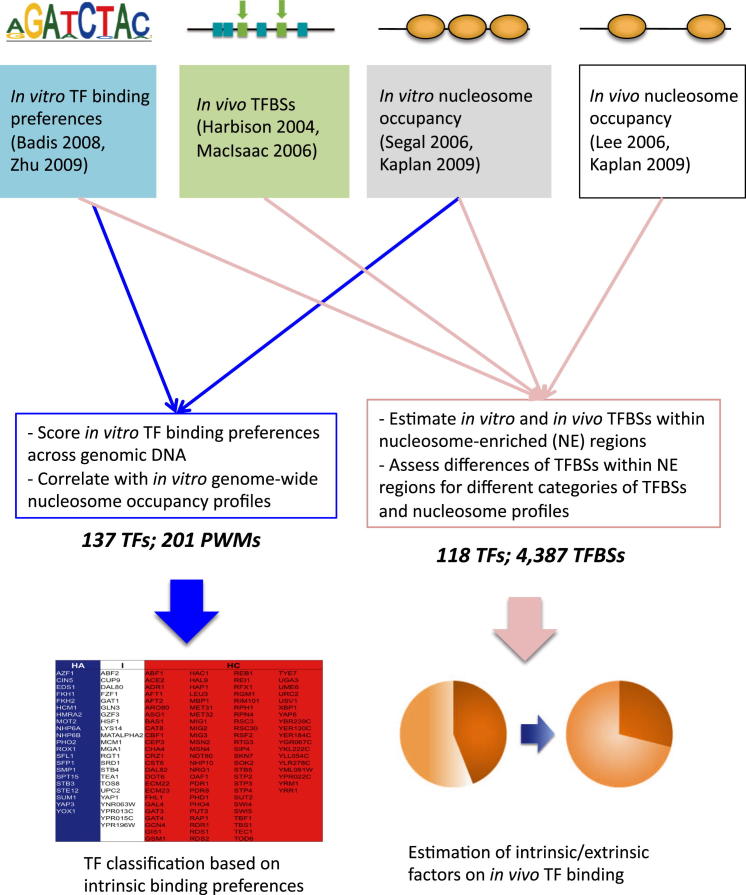
Data sets used in this study can be divided into four groups: (1) in vitro TF binding preferences from PBM experiments (Badis et al., 2008; Zhu et al., 2009), (2) in vivo TF binding sites from ChIP-chip (Harbison et al., 2004) (MacIsaac et al., 2006), and genome-wide nucleosome occupancy profiles determined (3) in vitro and (4) in vivo (Kaplan et al., 2009; Lee et al., 2007; Segal et al., 2006). In vitro TF binding preferences were used to score against the entire budding yeast genomic DNA. The predicted genome-wide TF binding preference landscapes were individually correlated against genome-wide nucleosome occupancy profile. We classified TFs into histone-correlated (HC), intermediate (I), and histone-anticorrelated (HA) groups according to these correlation coefficients (Figure 3 and Table 1). All four types of data sets were combined to compute the fractions of predicted and in vivo TFBSs likely to be occluded by nucleosomes, based on occupancy profiles in vitro and in vivo (Figure 5).