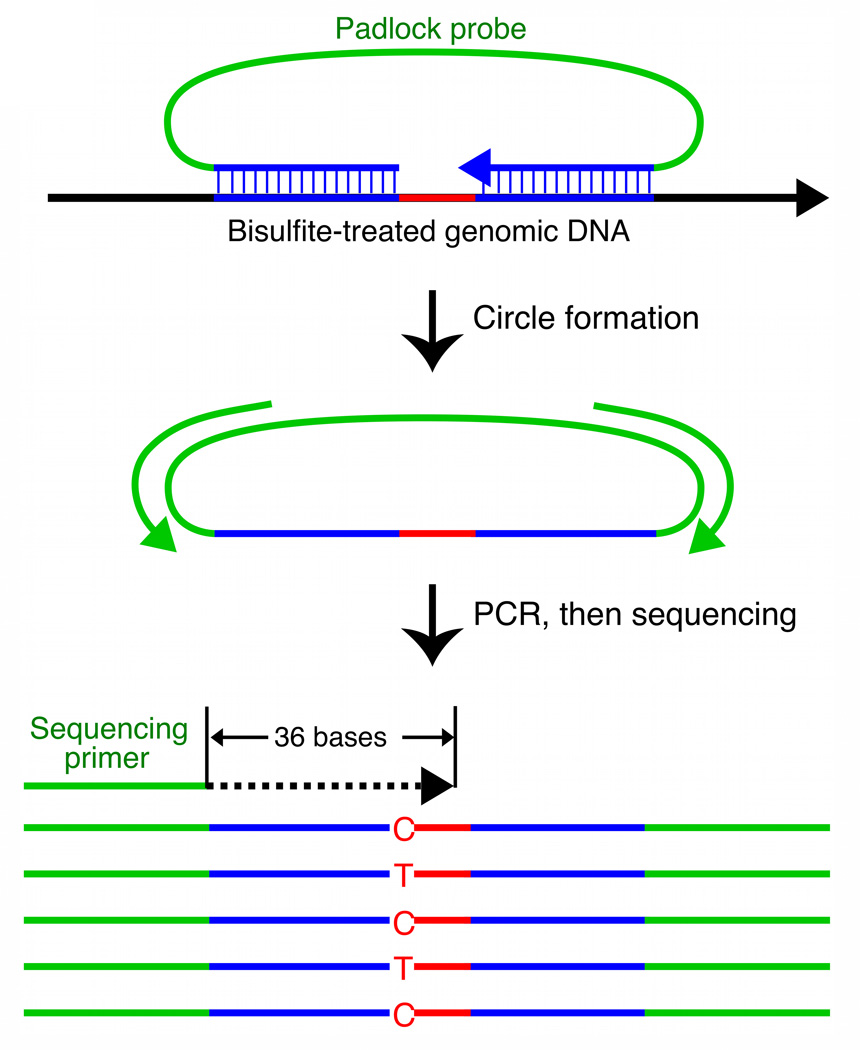
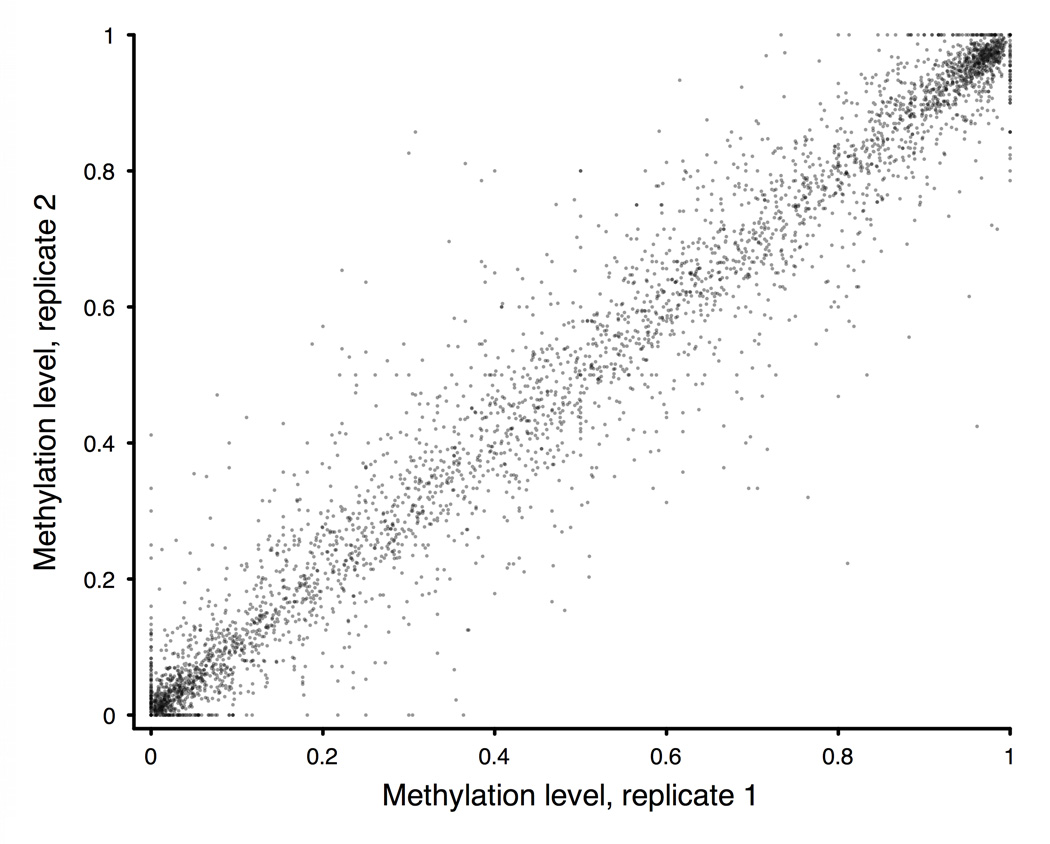
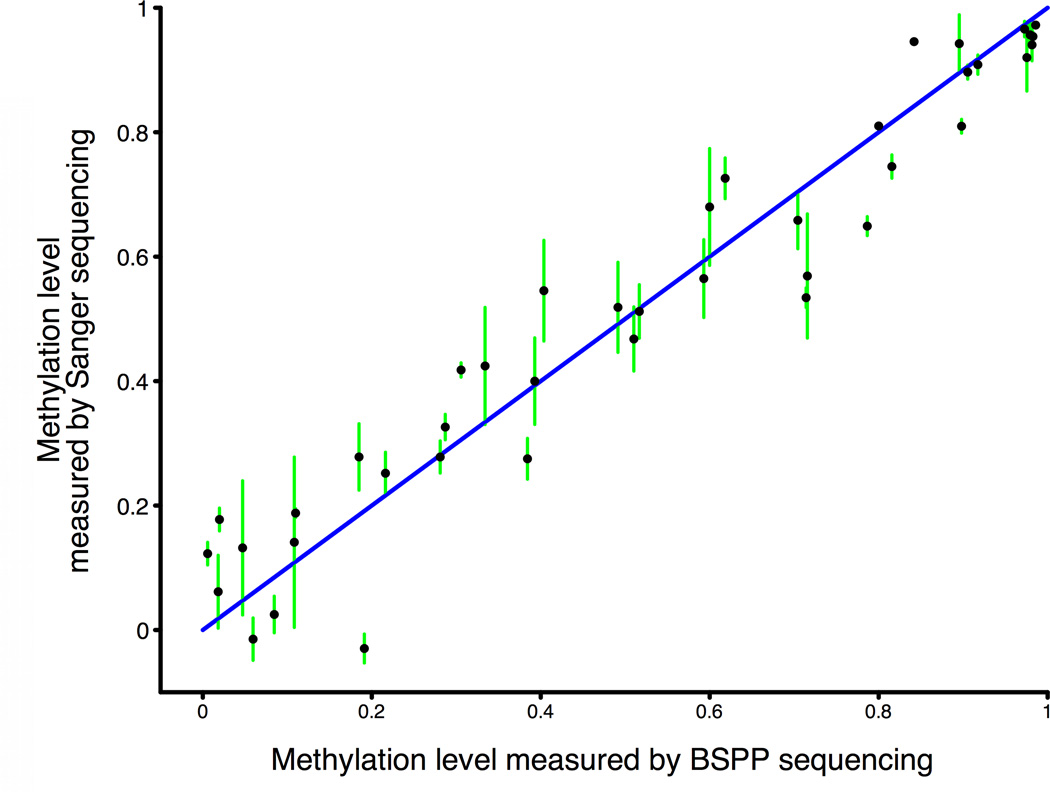
Figure 1. BSPP technology enabling accurate measurement of methylation levels.
a, BSPP experimental scheme. Two hybridizing locus-specific “arms” (blue) are connected by a 50bp common “backbone” sequence (green). In this work, ~10,000 BSPPs were designed to target CpG sites in bisulfite-treated DNA with a CpG located at the 3' end of the 10 bp polymerized span (red). Circles were formed by addition of polymerase, dNTP, and ligase, and were subsequently amplified using the backbone sequence as primers. Sequencing was then performed using an Illumina Genome Analyzer with a primer matching the backbone sequence; 28 bases of arm sequence were read through before sequencing informative positions within the span (read lengths were 36 bases in total). b, Correlation of methylation level in the technical replicates (Pearson coefficient r = 0.965). c, Correlation of BSPP methylation with the methylation levels determined by bisulfite PCR followed by Sanger sequencing at 33 locations (r = 0.966). Error bars (in green) represent the standard deviation of methylation as measured by Sanger sequencing.