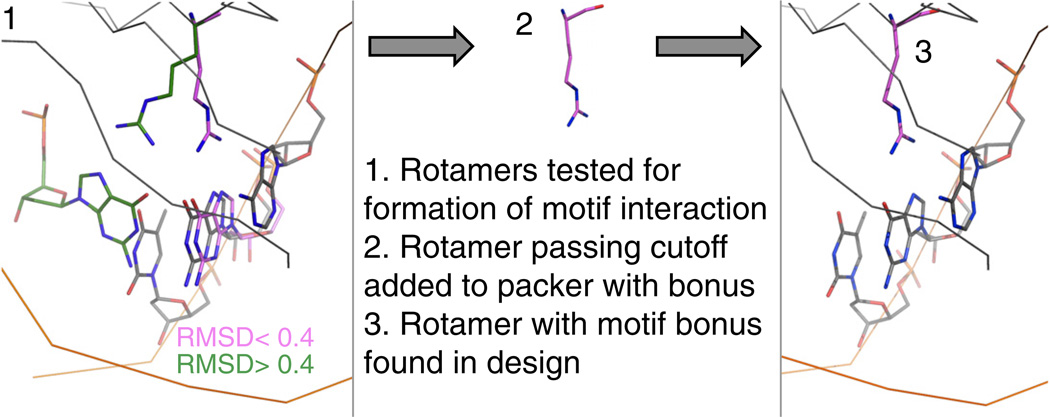
Fig. 2.
Overview of the motif-biased design protocol. In step 1, a series of rotamers and motifs are tested to see if they are compatible with the crystal structure undergoing design. These rotamers and motifs are subject to a series of cutoffs: distance of C1*, how parallel the placed base is to the crystal structure DNA, and RMSD of nucleobase atoms. In this example, two arginine rotamers (green and pink) are tested with a bidentate arginine–guanine motif, and the pink rotamer passes a nucleobase RMSD cutoff of < 0.4 when an ideal guanine base is placed in a motif-compatible position and compared to the nearest guanine base in the crystal structure. This pink arginine rotamer is then added to the standard rotamer sets used by the ROSETTA packer. The rotamer is given an energy bonus over other rotamers and is found in a design completed for this guanine base.