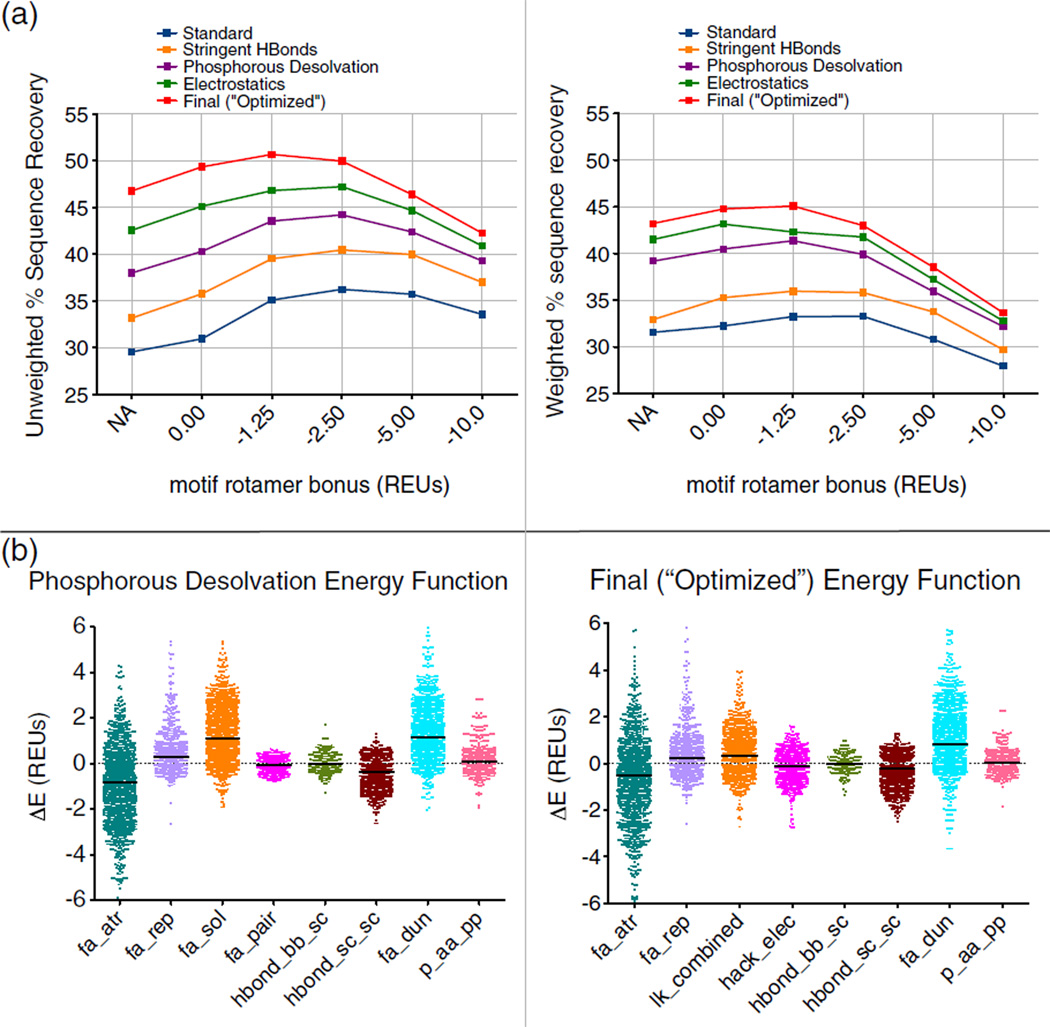
Fig. 3.
Optimization of ROSETTA energy function. Abbreviations for energy function terms are as follows: fa_atr, attractive; fa_rep, repulsive; fa_sol, solvation; fa_pair, distance-dependent atom pair potential; hbond_bb_sc, hydrogen bonds between backbone and side-chain atoms; hbond_sc_sc, hydrogen bonds between side-chain atoms; fa_dun, rotamer probability; p_aa_pp, probability of amino acid given backbone conformation; hack_elec, simple electrostatics; lk_combined, combination of terms for orientation-dependent desolvation model. (a) A comparison to two metrics of sequence recovery over several motif rotamer bonuses and several iterations of energy function optimization (Figs. S2–S5). The “Standard” energy function was the starting point for the optimization. The “Standard” energy function was improved by the addition of motifs, increasing the stringency of the hydrogen-bonding model (“Stringent HBonds”), modification of the phosphorous desolvation penalty (“Phosphorous Desolvation”), and the addition of a coulombic electrostatics term1 for the “Electrostatics” energy function. The “Final (“Optimized”)” energy function includes multiple additional changes detailed further in the text and in the supplement. (b) Energy differences, separated out by energy term, between incorrectly designed rotamers and rotamers with a motif bonus that match the native amino acid type, or more correctly match the native rotamer, than a designed rotamer with no bonus. The units for these energy differences are in REUs. The differences collected with the “Standard” energy function reveal that the solvation term (fa_sol) and the rotamer probability term26 (fa_dun) are the two energy terms that are being offset by the motif bonus. As a part of the energy function optimization, the solvation term was replaced with an orientation-dependent solvation model1 (lk_combined), and changes were made to the atom-specific desolvation parameters for several amino acid types.