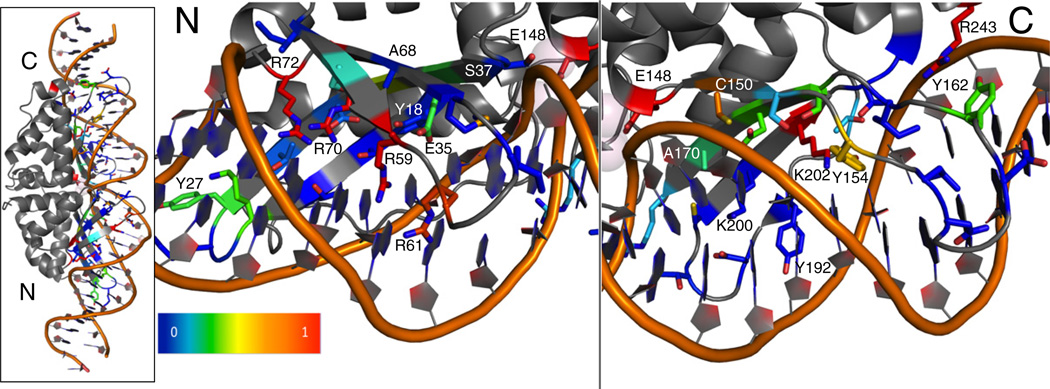
Fig. 5.
Visual representation of the interface conservation of I-AniI. The frequency of observing the wild-type amino acid after full randomization and selection (Fig. 4) is summarized on the structure of I-AniI. Only the 44 residues that were randomized are shown in this representation. Blue corresponds to a frequency of 0 or non-conserved positions. Red corresponds to positions that are highly conserved as the wild-type amino acid. The overall protein–DNA complex is shown on the leftmost panel, and the N- and C-terminal domains are separated in the other panels to allow for a closer examination of the conserved contacts. Four arginine residues are most conserved in the N-terminal domain and are likely essential for formation of the initial substrate-bound complex. Lys202 and Tyr154 are conserved in the C-terminal domain, and these interactions likely play an important role in the formation of the catalytic complex.18 This representation is incomplete in that it loses information if the preferred amino acid is not the wild type, but still a conserved type. For example, positions Tyr18, Tyr27, and Tyr162 are strongly conserved as aromatic residues (Fig. 4), but the native aromatic shows up at lower or equivalent frequencies as other aromatic types, resulting in blue or green shading at these positions.