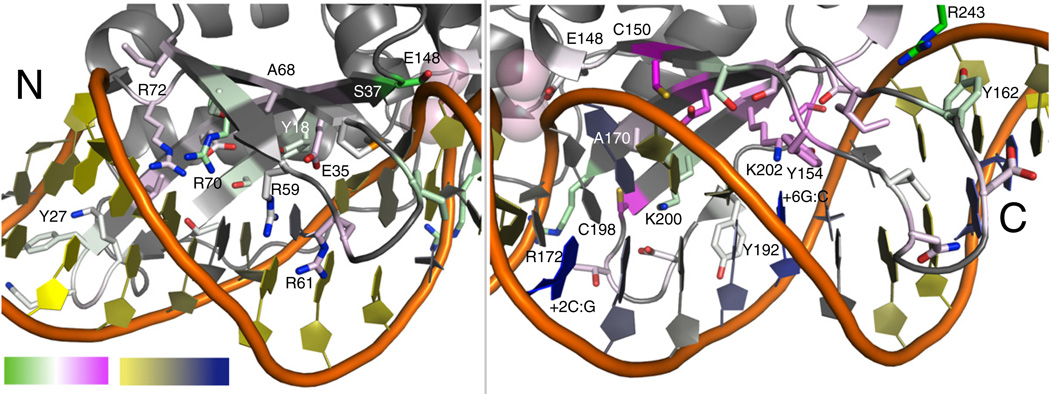
Fig. 7.
Recovery of experimental data with computational methods. A comparison between the two methods of sequence diversity generation, DNA-Rebuild and HighTemp-Packer, is summarized on the structure of I-AniI. The frequency distributions at each of the I-AniI interface positions were compared to the experimental data (Fig. 4) by both Euclidean distance and Jensen–Shannon divergence measures (Table 1 and Fig. S8). For this illustration, the Jensen–Shannon divergence measure33 calculated for the DNA-Rebuild method was subtracted from the same calculation completed for the HighTemp-Packer. White is designated as a value of 0, indicating that neither computational method better matched the experimental frequency distribution; green is negative values, indicating that the DNA-Rebuild performed better than the HighTemp-Packer; and pink is positive values, indicating that the HighTemp-Packer performed better than the DNA-Rebuild method. The DNA is colored based on the average RMSD between the DNA-Rebuild simulations and the crystal structure DNA, where yellow is the lowest average RMSD and where blue is the highest. The DNA moved farthest away from the crystal structure DNA in the same area that the DNA-Rebuild method performs well much less than the HighTemp-Packer, indicating that the DNA location has a significant effect on the design results.