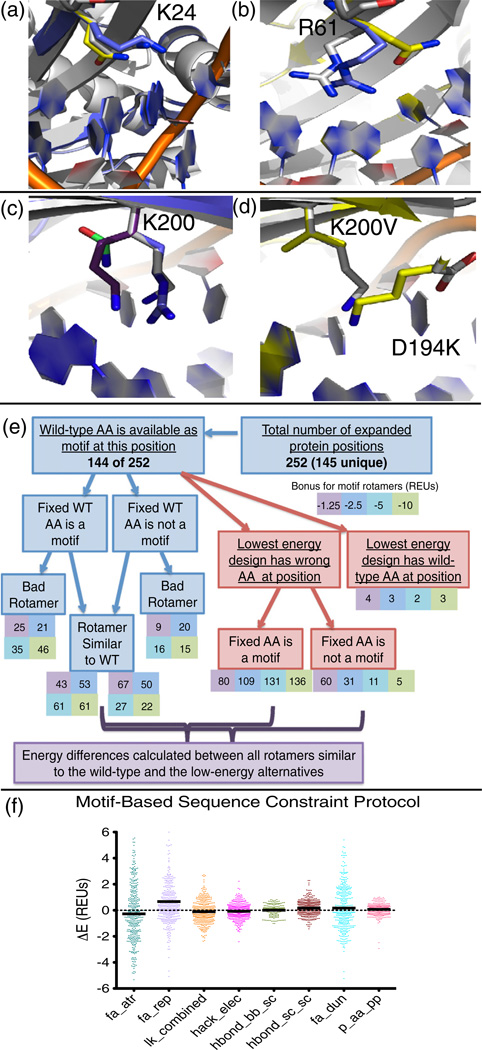
Fig. 8.
Motif-based sequence constraints. (a) Lys24 in the I-AniI interface (native rotamer, white) is mis-designed to a glutamine (yellow). The motif-based sequence constraint protocol revealed that position 24 can be a lysine motif, and the motif residue (blue) very closely matches the native lysine. (b) Arg61 in the I-AniI interface (native rotamer, white) is mis-designed to a glutamine (yellow). The motif-based sequence constraint protocol revealed that position 61 can be an arginine motif (blue). (c) The motif-based sequence constraint protocol showed that position Lys200 in the I-AniI interface (native rotamer, white) can be a motif of any of the three amino acid types previously identified to be active at this position (arginine, blue; lysine, purple; and asparagine, green). (d) The alternative low-energy design that disallows any of the motifs in (c) to be designed at position 200. The native structure is shown in white, and the design with K200V and D194K is shown in yellow. (e) Abbreviations: WT, wild type; AA, amino acid. Flowchart summarizing the results of the protocol that generates designs with forced amino acid types for each type of motif identified by the motif search. The protocol was completed only for protein positions that were considered to be true failures of the computational methods by a series of analyses. The chart summarizes the motif status, energetics, and rotameric state of the designs at each of these failed positions. Rotamers are considered similar to the wild-type amino acid if they have an RMSD of <0.8. (f) Energy differences calculated between rotamers that resemble the wild-type amino acid that has a motif rotamer incorporated with a bonus and between the incorrectly designed amino acid observed at this same protein position in the lowest-energy design, as marked on the flowchart in (e). The repulsive energy term (fa_rep) stands out at the biggest contributor to the energy difference between these rotamers.