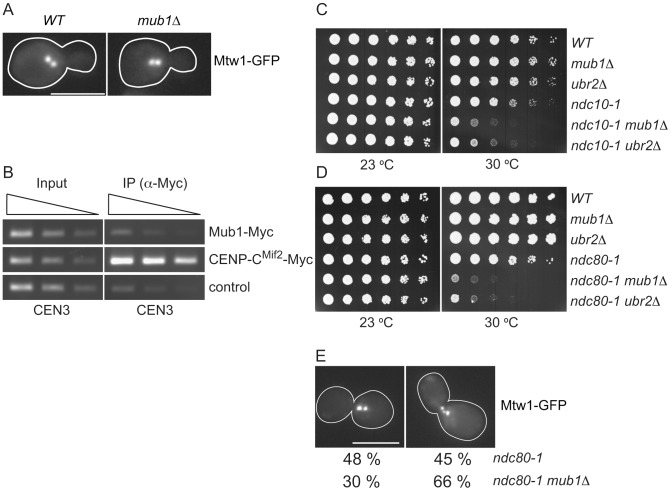
Figure 5. Mub1/Ubr2 cannot be detected at endogenous centromeres but are important for kinetochore function.
(A) Mub1 is dispensable for kinetochore biorientation. Mtw1-3GFP was monitored in either MUB1 (SBY3798) or mub1Δ (SBY8026) cells for biorientation defects. A representative cell from each strain is shown. Bar, 5 µm. (B) Mub1 cannot be detected at centromeres. Chromatin immunoprecipitation was carried-out on Mub1-Myc ubr2Δ (SBY8572), Mif2-Myc (SBY1566), and untagged control (SBY3) cells using a probe for CEN3. We obtained similar results using Mub1-Myc UBR2 cells (data not shown). (C, D) Mub1Δ and ubr2Δ exhibit negative genetic interactions with kinetochore mutants. Serial dilutions (5-fold) of ndc10-1 (SBY3, SBY7793, SBY7851, SBY164, SBY8613, SBY8773) and ndc80-1 (SBY3, SBY7793, SBY7851, SBY1117, SBY8436, SBY8432) kinetochore mutants with mub1Δ and ubr2Δ were plated on YPD and incubated at the indicated temperatures to examine genetic interactions. (E) Ndc80-1 mub1Δ double mutants exhibit an increase in declustered kinetochores. Ndc80-1 (SBY3934) and ndc80-1 mub1Δ (SBY8670) cells containing Mtw1-3GFP were released from G1 to 30 degrees and kinetochores were visualized at 180′. The percent of clustered (left panel) vs. unclustered (right panel) was quantified. Bar, 5 µm.