Abstract
Mutation of Arg427 and Arg472 in Rhizobium etli pyruvate carboxylase to serine or lysine greatly increased the activation constant (Ka) of acetyl CoA, with the increase being greater for the Arg472 mutants. These results indicate that while both these residues are involved in the binding of acetyl CoA to the enzyme, Arg472 is more important than Arg427. The mutations had substantially smaller effects on the kcat for pyruvate carboxylation. Part of the effects of the mutations was to increase the Km for MgATP and the Ka for activation by free Mg2+ determined at saturating acetyl CoA concentrations. The inhibitory effects of the mutations on the rates of the enzyme-catalysed bicarbonate-dependent ATP cleavage, carboxylation of biotin and phosphorylation of ADP by carbamoyl phosphate indicate that the major locus of the effects of the mutations was in the biotin carboxylase (BC) domain active site. Even though both Arg427 and Arg472 are distant from the BC domain active site, it is proposed that their contacts with other residues in the allosteric domain, either directly or through acetyl CoA, affect the positioning and orientation of the biotin-carboxyl carrier protein (BCCP) domain and thus the binding of biotin at the BC domain active site. Based on the kinetic analysis proposed here it is proposed that mutations of Arg427 and Arg472 perturb these contacts and consequently the binding of biotin at the BC domain active site. Inhibition of pyruvate carboxylation by the allosteric inhibitor, L-aspartate, was largely unaffected by the mutation of either Arg427 or Arg472.
Keywords: pyruvate carboxylase, allosteric regulator, site-directed mutagenesis, steady-state kinetics, sedimentation velocity
INTRODUCTION
Pyruvate carboxylase (PC, EC 6.4.1.1) is a key metabolic enzyme whose main function is the replenishment of tricarboxylic acid cycle intermediates that have been removed for synthetic purposes. In mammals, the anaplerotic role of PC is imperative to gluconeogenesis, adipogenesis, neurotransmitter synthesis and regulation of insulin release (1). PC is a biotin-dependent enzyme whose activity, in most organisms, is regulated by the allosteric activator, acetyl CoA. The degree of activation by acetyl CoA is different for PCs from different organisms, with the activity of some of them being totally independent of acetyl CoA whilst others are almost completely dependent on acetyl CoA for activity (2, 3). The activities of PC from most microbial sources, including Rhizobium etli PC (RePC; 4), are negatively regulated by L-aspartate, which acts as an antagonist to acetyl CoA activation (for a recent review see ref. 3).
Similar to other biotin-dependent carboxylases, the pyruvate carboxylation reaction is catalysed by PC in three steps (Fig. 1). Briefly, bicarbonate is initially activated in the biotin carboxylase (BC) domain via phosphorylation by MgATP to form the putative carboxyphosphate intermediate (reaction i). The covalently attached biotin prosthetic group is then carboxylated in the BC domain to form carboxybiotin in reaction (ii). The reversible decomposition of the carboxyphosphate intermediate results in the formation of Pi and CO2. Pi most likely acts as the active site base, deprotonating the covalently attached biotin at the N1-position, while CO2 carboxylates the resulting biotin-enolate. Carboxybiotin is then translocated to the carboxyl transferase (CT) domain where the carboxyl group is transferred from carboxybiotin to pyruvate, producing oxaloacetate (reaction iii) (5–7). The biotin carboxyl carrier protein (BCCP) domain, to which the biotin prosthetic group is covalently attached, facilitates the intersubunit movement of carboxybiotin from the BC domain to the CT domain of an adjacent subunit.
Figure 1.

PC catalyses the reaction of pyruvate carboxylation in two steps. Reactions (i) and (ii) occur in the BC domain of the enzyme and result in the carboxylation of the covalently attached biotin. Reaction (iii) occurs in the CT domain and results in the formation of oxaloacetate via the decarboxylation of carboxybiotin and concomitant carboxylation of pyruvate.
Acetyl CoA was shown to have a major stimulatory effect on reactions (i) and (ii) (8–10) and only a small effect on reaction (iii) (10, 11). The steady-state rates of ATP cleavage (8–10) and the carboxylation of free biotin (12) are stimulated by acetyl CoA, while the rate of the approach to steady-state for the formation of enzyme-carboxybiotin complex was also dependent on acetyl CoA concentration (9). Attwood and Graneri (8) and Zeczycki et al. (10) also showed an interrelationship between the activation of the bicarbonate-dependent ATP cleavage and pyruvate carboxylation reactions by acetyl CoA and Mg2+. Using chimeric enzymes generated from the two yeast isoforms of PC (Pyc1 and Pyc2), Jitrapakdee et al. (13) showed that the extent of the activation of the enzyme by acetyl CoA was largely determined by the BC domain.
Recently determined structures of RePC (14, 15), human PC and Staphylococcus aureus PC (16–18) have led to an improved molecular-level description of PC function. In particular, the high-resolution X-ray crystal structure of RePC complexed with ethyl CoA, a non-hydrolysable analogue of acetyl CoA, in the allosteric domain has disclosed several residues poised to play a key role in the binding of acetyl CoA. The acetyl CoA binding site lies at the interface between the BC domain and a distinct allosteric domain which physically links the BC, CT and BCCP domains (Fig 2a). A detailed view of the allosteric binding site of RePC highlights the interactions between the guanidinyl groups of Arg427 and Arg472 and the 3′-phosphate and 5′-α-phosphate, respectively, of ethyl CoA (Fig 2b). The positioning of Arg427 and Arg472 relative to ethyl CoA suggests that these residues may be important for the proper binding and orientation of acetyl CoA in the allosteric domain and, thus, to the overall allosteric regulation of the enzymatic activities.
Figure 2.
(a) The allosteric binding site (green), with ethyl CoA bound, lies in part between the BC (blue) and CT (yellow) domains and between the CT and BCCP (red) domains. (b) Detailed view of the interactions between nucleotide portion of ethyl CoA and residues contained in BC and allosteric domains, including Arg427 and Arg472. Reproduced from St. Maurice et al. (2007).
In this study, we have performed site-directed mutagenesis of residues Arg427 and Arg472 to produce four mutants: R427S, R427K, R472S, and R472K. We have performed kinetic analyses of these mutants to investigate the roles of residues Arg427 and Arg472 in binding the allosteric activator and the effects on the induction of allosteric changes that lead to enhancement of catalysis. In addition, we have examined the effects of these mutations on the allosteric inhibition of RePC by L-aspartate.
MATERIALS AND METHODS
Materials
IPTG, malate dehydrogenase and lactate dehydrogenase were obtained from Roche. HisPur™ cobalt IMAC resin was obtained from Thermo Scientific. All other materials were purchased from Sigma-Aldrich
Construction of the RePC mutants
Mutagenesis was conducted on the 1.0 kb Xho I – Sac II PC gene fragment corresponding to the allosteric domain using a Quickchange site-directed mutagenesis kit (Stratagene). Mutations were verified by DNA sequencing (Macrogen, Korea). The primers used to generate R427S, R427K, R472S, and R472K are listed in the Table S1 (Supporting Information). The equivalent fragment of the wild-type RePC gene in the expression clone (14, 19) was then replaced with the mutagenized fragments.
Expression and purification of RePC
The bacteria Escherichia coli BL21-(DE3), containing the pCY216 plasmid (20), which encodes the E. coli BirA gene, were transformed with either the wild-type RePC or mutant RePC plasmid. The cultures were grown in 8 L Luria-Bertani broth supplemented with 6.25 g/L arabinose, 10 mg/L biotin, 200 mg/L ampicillin, and 30 mg/L chloramphenicol at 37 °C until an OD600 of 1.0–1.2 was reached. The cultures were subsequently cooled on ice for 30 min, induced with addition of 0.1 mM IPTG, and incubated for approximately 36 hours at 16 °C. The cells were harvested by centrifugation at 4039g and 4 °C for 15 min. The harvested cells were disrupted by incubation with 1 mg/mL lysozyme followed by mechanical disruption of the cells using a Bead-Beater (Biospec). Nucleic acids were removed from the lysate by protamine sulphate precipitation and the total proteins were subsequently precipitated with 36% (w/v) saturated ammonium sulphate. The total proteins were then suspended in loading buffer (300 mM NaCl, 50 mM NaH2PO3, and 10 mM imidazole, pH 7.4) prior to loading onto a 20 ml of HisPur™ cobalt resin. RePC was selectively eluted from the resin using elution buffer (300 mM NaCl, 50 mM NaH2PO3, and 150 mM imidazole, pH 7.4). Purified PC was stored at −80 °C in storage buffer containing 30% (v/v) glycerol, 0.1 M Tris-HCl (pH 7.8), and 1 mM DTE (12).
Determination of the biotin content of RePC
Aliquots of the enzyme were digested in triplicate with 0.2% (w/v) chymotrypsin (Sigma) in 0.2 M KH2PO4 (pH7.2) at 37 °C for 24 hours followed by digestion with 0.45% (w/v) protease from Streptomyces griseus at 37 °C for 48 hours. The biotin assay was performed as described by Rylatt et al. (21) in triplicate. The enzyme concentrations referred to are determined by the total amount of enzymic biotin in the purified wild-type and mutant RePC proteins.
Sedimentation analysis of the enzymic quaternary structure
Sedimentation velocity analytical centrifugation was performed with a Beckman Proteome Lab XL-A (Beckman-Coulter, Palo Alto, CA) ultracentrifuge using the absorbance optics system to visualize the protein at a wavelength of 280 nm. Two-sector cells were used and data were acquired at every 0.003 cm. Data were collected as 300 absorbance scans with a nominal time increment of 1 min at 30 °C and at a speed of 40000 rpm. In all cases enzyme samples were prepared at a concentration of 2 μM in 0.1 M Tris-HCl (pH7.8), 20 mM NaHCO3, 5 mM MgCl2, 10 mM pyruvate, 0.1 mM acetyl CoA, and 1 mM DTE. The computer-captured data were analysed with SEDFIT (22). The partial specific volume of the enzyme was calculated from the amino acid composition using SEDNTERP [www.bbri.org/RASMB] (22). The density of the Tris-HCl buffer (1.005 g/mL) was assumed to be the density of the enzyme solution.
Pyruvate carboxylation activity assays
The initial rates of the enzymatic carboxylation of pyruvate were determined using a coupled spectrophotometric assay in which the oxaloacetate was converted to malate using malate dehydrogenase. The concomitant oxidation of NADH was measured by the change in absorbance at 340 nm (23). The enzymic activity was determined at 30 °C in a 1 mL reaction mixture containing 0.1 M Tris-HCl (pH 7.8), 6 mM MgCl2, 20 mM NaHCO3, 10 mM pyruvate, 0.22 mM NADH and 5 units of malate dehydrogenase. In reactions where the concentration of acetyl CoA was varied, 1 mM MgATP was present in all assays. In the assays where MgATP concentrations were varied, reactions with the wild-type enzyme contained 0.25 mM acetyl CoA. Assays to determine mutant RePC activity were performed at acetyl CoA concentrations of 0.5 mM (R427S), 2 mM (R427K) or 5 mM (R472S and R472K). These acetyl CoA concentrations were also used in the experiments where free Mg2+ concentrations were varied at a constant MgATP concentration (1 mM). Inhibition of pyruvate carboxylation activity was determined in the presence of 1 mM MgATP at varied concentrations of L-aspartate (0 – 60 mM) in the absence of acetyl CoA. Assays were initiated with the addition of enzyme (final concentrations 0.02 – 0.2 μM). Apparent kcat values were calculated by dividing the measured reaction velocity by the biotin concentration of the RePC used in the assay.
Bicarbonate-dependent ATP-cleavage activity assays
The specific activities for the wild-type and mutant RePC-catalysed bicarbonate-dependent ATP-cleavage were determined in triplicate using a coupled spectrophotometric assay where pyruvate kinase and lactate dehydrogenase were used as coupling enzymes (8). The reactions were performed at 30° C in a 1 mL reaction volume containing 0.1 M Tris-HCl (pH 7.8), 5 mM MgCl2, 1 mM MgATP, 20 mM NaHCO3, 10 mM phosphoenolpyruvate, 0.22 mM NADH, 5 units of pyruvate kinase and 4 units of lactate dehydrogenase. Activities were determined both in the presence or absence of saturating concentrations of acetyl CoA (0.25 mM for the wild-type enzyme, 0.5 mM for R427S, 2 mM for R427K, 5 mM for R472S and R472K mutant RePC enzymes) and initiated with the addition of enzyme (final concentration 2 μM.)
Phosphorylation of MgADP by carbamoyl phosphate
The rate of ADP phosphorylation by carbamoyl phosphate was determined for the wild-type and RePC mutant enzymes in triplicate, using a spectrophotometric assay where hexokinase and glucose-6-phosphate dehydrogenase were used as coupling enzymes (24). Reactions were performed at 30° C in a 1 mL reaction mixture containing 0.1 M Tris-HCl (pH 7.8), 8 mM MgCl2, 2 mM ADP, 10 mM carbamoyl phosphate, 0.5 mM glucose, 0.5 mM NADP, 5 units of hexokinase, 4 units of glucose-6-phosphate dehydrogenase, in the presence or absence of saturating concentrations of acetyl CoA (0.25 mM for the wild-type enzyme, 0.5 mM for R427S, 2 mM for R427K, 5 mM for R472S and R472K). Reactions were initiated with the addition of enzyme (concentration of 2 μM).
Carboxylation of free biotin
The rate of free biotin carboxylation was performed essentially as described by Adina-Zada et al. (12). The reaction mixture contained 0.1 M Tris-HCl (pH 7.8), 10 mM biotin, 1 mM MgATP, 5 mM MgCl2, 20 mM NaHCO3, 10 μCi/mL NaH14CO3, and saturating concentrations of acetyl CoA (0.25 mM for the wild-type enzyme, 0.5 mM for R427S, 2 mM for R427K, 5 mM for R472S and R472K RePC mutant enzymes). Reactions were performed at 30° C and were initiated with the addition of enzyme to a final concentration of either 0.5 μM (wild-type enzyme) or 10 μM (RePC mutant enzymes). At one-minute intervals over a period of six minutes, the reaction was terminated by a rapid transfer (in triplicate) of 0.1 mL aliquots to 0.9 ml cold denaturating solution, (water and n-octanol; 35:1), chilled on ice. The remaining H14CO3−/14CO2 was removed by bubbling CO2 gas through the solutions for 40 min at room temperature. 0.5 mL aliquots of gassed solutions were subsequently added to scintillation fluid and the radioactivity due to the presence of 14C-carboxybiotin was counted. Controls were performed in which aliquots of the reaction mixture were transferred to the termination solution before the initiation of the reaction. After enzyme had been added to these quenched aliquots, CO2 was bubbled through as above and the radioactivity was determined. The endogenous radioactivity in these control samples was subtracted from that determined in the enzymatic reactions. Specific radioactivity of NaH14CO3 was determined by measuring the radioactivity of aliquots of a reaction mixture containing a known number of moles of total NaHCO3. The rates of biotin carboxylation were calculated by linear regression analysis of the reaction time-courses.
Data analysis
The dependence of pyruvate carboxylation activity on acetyl CoA concentration was analysed using nonlinear least-squares regression fits of the initial velocities determined at varying acetyl CoA concentrations to equation (1)
| (1) |
where [A] is the concentration of acetyl CoA, Ka is the activation constant and h is the Hill coefficient of cooperativity. The kappcat is the apparent rate constant at each concentration of acetyl CoA, k°cat is the catalytic rate constant of the acetyl CoA-independent reaction and kcat is the catalytic rate constant of the acetyl CoA-dependent reaction.
The dependence of pyruvate carboxylation activity on the concentration of free Mg2+, which is treated as a pseudo-substrate in this kinetic examination, was analysed using nonlinear least-squares regression fits of the initial velocities determined at varying Mg2+ concentrations ([A]) to the Michaelis-Menten (equation 2).
| (2) |
Inhibition of pyruvate carboxylation by L-aspartate was analysed by non-linear regression fits of the data to equation (3)
| (3) |
where the total activity is expressed as a percentage of the activity in the absence of L-aspartate. R is the pyruvate carboxylation activity in the presence of saturating concentrations of L-aspartate which is expressed as a percentage of the activity in the absence of L-aspartate. Ki is the apparent inhibition constant and [Asp] is the concentration of L-aspartate.
RESULTS
Effects of mutations at Arg427 and Arg472 on the quaternary structure of RePC
Sedimentation velocity analytical ultracentrifugation was used to determine if the incorporation of mutations at Arg427 and Arg472 resulted in the destabilisation of the overall tetrameric arrangement of RePC (Fig. 3). With the exception of the R472K mutant, the wild-type and mutant RePC enzymes predominantly existed (> 70%) in the tetrameric form under conditions that closely mimicked those used for the steady-state kinetic analysis (Table 1). The R472S mutant does display a more highly resolved monomer peak in the sedimentation profile than the wild-type and R427K/S mutants, but still retains more than 70% of the tetrameric form of the enzyme. Based on the sedimentation profile, approximately 45% of the R472K enzyme was estimated to exist as a tetramer in the presence of 0.1 mM acetyl CoA. The effect of higher concentrations of the activator on the quaternary structure of the R427K mutant could not be determined since the increased absorbance, resulting from increasing concentrations of acetyl CoA, did not allow for the accurate measurement of the protein absorbance.
Figure 3.
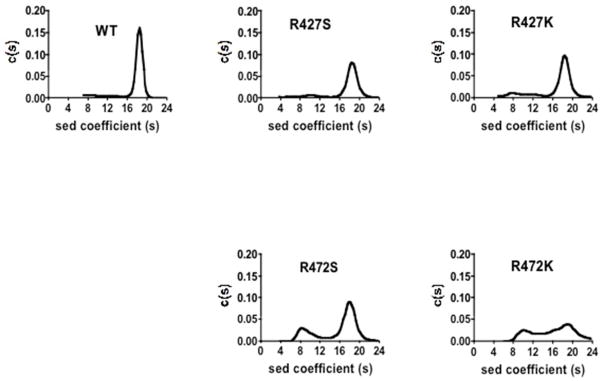
Sedimentation velocity analyses of wild-type RePC and the R427S, R427K, R472S, R472K RePC mutants. Experiments were performed as described in Materials and Methods. Plots of derived sedimentation coefficients distribution [c(s)] vs sedimentation coefficient obtained from the application SEDFIT and maximum entropy method of analysis to the original apparent sedimentation coefficient distribution vs apparent sedimentation coefficient data (g(s*) vs S8 plots).
Table 1.
Molecular masses and quaternary structure compositions of wild-type, R427S, R427K, R472S, and R472K determined by analytical ultracentrifugation (see Figure 2)
| RePC Enzyme | Monomer
|
Dimer
|
Tetramer
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Sedimentation coefficient | Molecular mass (kDa) | % | Sedimentation coefficient | Molecular mass (kDa) | % | Sedimentation coefficient | Molecular mass (kDa) | % | |
| WT | Mixture of monomers and dimers | 16 | 18.34 ± 0.80 | 525 ± 31 | 84 | ||||
| R427S | 5.89 ± 0.79 | 105 ±20 | 4 | 10.50 ± 1.79 | 255 ± 63 | 13 | 18.52 ± 1.34 | 589 ± 67 | 83 |
| R427K | 7.70 ± 1.23 | 145 ± 33 | 13 | 12.26 ± 1.40 | 293 ± 48 | 12 | 17.9 ± 1.51 | 507 ± 64 | 75 |
| R472S | 9.25 ± 1.56 | 195 ± 51 | 28 | - | - | - | 17.9 ± 1.51 | 507 ± 64 | 72 |
| R472K | Mixture of tetramers, dimers and monomers | ||||||||
Analysis performed in 0.1 M Tris-HCl (pH7.8), 20 mM NaHCO3, 5 mM MgCl2, 10 mM pyruvate, 0.1 mM acetyl CoA, and 1 mM DTE with 2 μM enzyme.
Effects of mutations at Arg427 and Arg472 on the binding of acetyl CoA and the activation of the pyruvate carboxylation reaction
In order to investigate the effects of the mutations on the overall reaction catalysed by RePC, the pyruvate carboxylation activities for each of the RePC mutants were determined at fixed concentrations of substrates, that were saturating for the wild-type RePC, and varying concentrations of acetyl-CoA (Fig. 4). Preliminary data for the effect of the R472S mutation on the Ka for acetyl CoA was reported by Adina-Zada et al. (25). The estimates of the kinetic parameters derived from fits of the initial rates of pyruvate carboxylation to eqn (1) are presented in Table 2. The value of Ka for acetyl CoA was 15-fold, 76-fold, 203-fold and 252-fold greater than that determined for the wild-type enzyme in the R427S, R427K, R472S and R472K mutant enzymes, respectively. The increase in Ka indicates that these mutations have greatly reduced the affinity of the enzyme for acetyl CoA. Further, the reduced apparent kcat values for the four mutants (R427S, R427K, R472S and R472K were 41%, 32%, 25% and 32% of the wild-type enzyme kcat) suggests that the mutations reduced the ability of acetyl CoA to stimulate catalysis, even at saturating concentrations. Values of k°cat were approximately 2- and 6-fold greater in the R427S- and R427K- catalysed reactions, respectively, than that determined for the wild-type enzyme. These results indicate that mutations at Arg427 had a positive effect on the acetyl CoA-independent pyruvate carboxylation activity. In contrast, the kcat values determined for the Arg472 mutant-catalysed reactions were not significantly different from that of the wild-type catalysed reaction (t-test; p > 0.2). The Hill coefficients of cooperativity that were determined ranged from 2.3 and 3.0 for all forms of the enzyme except for the R427K mutant, which had a Hill coefficient of 1.3, suggesting a marked reduction in the cooperative activation of this mutant by acetyl CoA.
Figure 4.

Activation of the wild-type RePC and R427S, R427K, R472S, R472K RePC mutant catalysed carboxylation of pyruvate by acetyl CoA. Solid lines represent nonlinear least-squares regression fits of the data to equation (1). Preliminary data for R427S was reported by Adina-Zada et al. (2011).
Table 2.
Kinetic parameters for pyruvate carboxylation with varied concentrations of acetyl CoA
| Enzyme | k°cat(s−1) | kcat (s−1) | Ka acetyl CoA (μM) | h |
|---|---|---|---|---|
| WT | 0.15 ± 0.01 | 10.6 ± 0.2 | 8.1 ± 0.2 | 2.8 ± 0.2 |
| R427S | 0.26 ± 0.03 | 4.3 ± 0.1 | 121 ± 2 | 3.0 ± 0.1 |
| R427K | 0.67 ± 0.03 | 3.4 ± 0.2 | 614 ± 50 | 1.3 ± 0.1 |
| R472S | 0.10 ± 0.04 | 2.6 ± 0.1 | 1640 ± 60 | 2.4 ± 0.2 |
| R472K | 0.17 ± 0.04 | 3.4 ± 0.1 | 2040 ± 75 | 2.3 ± 0.2 |
Assay conditions: 100 mM Tris-HCl (pH 7.8), 30°C, 20 mM NaHCO3, 1 mM MgATP, 5 mM MgCl2, acetyl CoA (0 – 6 mM). The reported errors are standard errors of the values of the parameters calculated from the nonlinear regression fits of the data shown in Fig. 2 to equation (1).
Pyruvate carboxylation activity was also measured as a function of MgATP concentration in the presence of saturating acetyl CoA (Fig. 5, Table 3). The kcat values for the wild-type and mutant enzyme-catalysed reactions were similar to those determined when acetyl CoA was varied (see above), but the mutants exhibited higher Km values for MgATP relative to the wild-type enzyme (1.5-fold; 2.6-fold; 8.7-fold and 6.7-fold greater for R427S, R427K, R472S and R427K respectively). The combined effects of the mutations on kcat and the Km result in kcat/Km values that are 26%, 12%, 2% and 3%, respectively, of the wild-type enzyme. The smaller values of kcat/Km for the Arg472 mutants indicate that mutation of this residue has a larger effect on reaction steps from MgATP binding to the first irreversible step in the reaction than mutation of Arg427 does.
Figure 5.
The dependence of apparent kcat for the pyruvate carboxylation reaction catalysed by wild-type RePC (■), R427S (●), R427K (○), R472S (▲), and R472K (▽) mutant RePC on the concentration of MgATP. Solid lines represent nonlinear least-squares regression fits of the data to Michaelis-Menten equation.
Table 3.
Kinetic parameters for pyruvate carboxylation with varied concentrations of MgATP
| Enzyme | kcat (s−1) | Km (mM) | kcat/Km (mM−1s−1) |
|---|---|---|---|
| WT | 10.9 ± 0.3 | 0.11 ± 0.01 | 99 |
| R427S | 4.2 ± 0.2 | 0.16 ± 0.02 | 26 |
| R427K | 3.4 ± 0.2 | 0.29 ± 0.04 | 12 |
| R472S | 2.0 ± 0.2 | 1.0 ± 0.2 | 2 |
| R472K | 1.9 ± 0.2 | 0.7 ± 0.2 | 3 |
Assay conditions: 100 mM Tris-HCl (pH7.8), 30°C, 20 mM NaHCO3, 5 mM MgCl2, saturated concentrations of acetyl CoA (see Materials and Methods), and 0.02 – 1.5 mM MgATP. The reported errors are standard errors of the parameters calculated from the nonlinear regression fit of the data to the Michaelis-Menten equation.
The effect of mutations at Arg427 and Arg472 on the activation of pyruvate carboxylation by free Mg2+
Fig. 6 and Table 4 show the effect of free Mg2+ on the initial rates of the wild-type and Arg427/Arg472 RePC mutant catalysed carboxylation of pyruvate in the presence of saturating concentrations of acetyl CoA. Mutations of Arg427 have small effects on the Ka for Mg2+, with observed Ka values that were 2-fold and 8-fold (R427S and R427K, respectively) greater than the wild-type Ka. The mutation of Arg472 has a more pronounced effect on the Ka for Mg2+ with 30- and 37-fold increases (R472S and R472K, respectively), relative to the wild-type enzyme.
Figure 6.
Activation of the wild-type RePC (■), R427S (●), R427K (○), R472S (▲), and R472K (▽) RePC mutant-catalysed carboxylation of pyruvate by Mg2+ in the presence of saturating concentrations of acetyl CoA. Solid lines represent nonlinear regression fits of the data to equation (2).
Table 4.
Mg2+ activation of the pyruvate carboxylation reaction
| Enzyme | Ka Mg2+ (mM) |
|---|---|
| WT | 0.11 ± 0.02 |
| R427S | 0.20 ± 0.04 |
| R427K | 0.90 ± 0.22 |
| R472S | 3.3 ± 0.8 |
| R472K | 4.1 ± 1.0 |
Assay conditions: 100 mM Tris-HCl (pH7.8), 30°C, 20 mM NaHCO3, 1 mM MgATP, saturating concentrations of acetyl CoA (see Materials and Methods), and 0.115 – 12 mM free Mg2+. The reported errors are standard errors of the values of the parameters estimated from the nonlinear regression fits of the data shown in Fig. 6 to equation (2).
The effect of mutations at Arg427 and Arg472 on the reactions that are catalysed in the BC domain of RePC
A. Bicarbonate-dependent ATP cleavage
The activity of the bicarbonate-dependent ATP cleavage reaction was determined for the wild-type and mutant RePC enzymes in the presence and absence of saturating concentrations of acetyl CoA. As shown in Table 5, in the presence of acetyl-CoA, the ATP-cleavage activities of R427S, R427K, R472S and R472K are 1.1%, 0.4%, 0.6% and 0.6%, respectively, of the wild-type enzyme activity. In the absence of acetyl-CoA, the bicarbonate-dependent ATPase activities of R427S, R427K, R472S and R472K are 5.8%, 0.8%, 1.2%, 0.9%, respectively, of the activity of wild-type enzyme activity.
B. Phosphorylation of ADP by carbamoyl phosphate
Using carbamoyl phosphate, a stable substrate analogue of carboxyphosphate, the effects of the Arg427 and Arg472 mutations on the partial reverse reaction of the BC domain can be assessed by measuring the catalytic formation of MgATP (24)]. To determine whether the mutations of Arg427 and Arg472 affect this part of the reaction in the BC domain active site, the rates of ADP phosphorylation by carbamoyl phosphate was determined for all the mutant enzyme forms in the presence and absence of acetyl CoA (Table 6). In the presence of acetyl CoA, the activities of R427S, R427K, R472S and R472K were determined to be 17%, 21%, 5%, 8%, respectively, of the wild-type enzyme activity. In the absence of acetyl CoA, both the wild-type and R427S enzyme showed a similar reduction in ADP phosphorylation activity compared to that determined in the presence of acetyl CoA. Interestingly, the R427K, R472S and R472K mutants all exhibited a slightly increased activity in the absence of acetyl CoA.
Table 6.
Effects of acetyl CoA on rates of MgADP phosphorylation by carbamoyl phosphate
| Enzyme | kcat (s−1 ) (saturating acetyl CoA) | % | kcat (s−1) (no acetyl CoA) | % |
|---|---|---|---|---|
| WT | 0.27 ± 0.01 | 100 | 0.19 ± 0.01 | 100 |
| R427S | 0.048 ± 0.002 | 17 | 0.032 ± 0.003 | 17 |
| R427K | 0.056 ± 0.006 | 21 | 0.077 ± 0.008 | 41 |
| R472S | 0.015 ± 0.001 | 5 | 0.026 ± 0.001 | 14 |
| R472K | 0.021 ± 0.001 | 8 | 0.039 ± 0.002 | 21 |
Assay conditions: 100 mM Tris-HCl (pH7.8), 30°C, 8 mM MgCl2, 2 mM ADP, 10 mM carbamoyl phosphate, saturating concentrations of acetyl CoA (see Materials and Methods). The reported errors are standard deviation of the means of three separate determinations of the apparent kcat values.
C. Carboxylation of free biotin
The steady-state measurement of ATP-cleavage involves both ATP-cleavage and the carboxylation of biotin. To further clarify the role of acetyl CoA in the BC domain reaction, the rate of free biotin carboxylation was measured for the wild-type enzyme and the mutants (Table 7). The mutants exhibited rates of biotin carboxylation that were 2% –6% of the wild-type enzymatic activity. The rates for the Arg472 mutants were 30–50% of those determined with the Arg427 mutants. These results indicate that the incorporated mutations have reduced the ability of the enzyme to form carboxybiotin, with mutations at Arg472 having significantly larger effects than mutation of Arg427 (p <0.01 in paired t-tests).
Table 7.
Rate of biotin carboxylation
| Enzyme | kcat (s−1 ) | % |
|---|---|---|
| WT | 1.47±0.03 | 100 |
| R427S | 0.094±0.001 | 6 |
| R427K | 0.081±0.001 | 6 |
| R472S | 0.034±0.001 | 2 |
| R472K | 0.046±0.001 | 3 |
Assay conditions: 100 mM Tris-HCl (pH7.8), 30°C, 20 mM NaHCO3, 10 μCi/mL NaH14CO3, 6 mM MgCl2, 1 mM ATP, 10 mM biotin, saturating concentrations of acetyl CoA (see Materials and Methods). The reported errors are the standard errors of the estimates of the kcat from the linear regression analysis of the time-courses of carboxybiotin formation.
The effect of mutations at Arg427 and Arg472 on the inhibition of pyruvate carboxylation by L-aspartate
The effects of mutations at Arg427 and Arg472 on the inhibition of RePC-catalysed carboxylation of pyruvate by L-aspartate in the absence of acetyl CoA were examined (Fig 7, Table 8). The Arg427 and Arg472 mutations had little effect on the apparent Ki for L-Aspartate (values of Ki for the mutants were not significantly different from that of the wild-type enzyme – t-test; p > 0.2). Similarly, the residual activities of the mutants were not significantly different from that determined with the wild-type enzyme (t-test; p > 0.2), with the exception of the R427K mutant. The residual activity at saturating concentrations of L-aspartate for the R427K-catalysed reaction was determined to be 2-fold greater than the residual activity of the wild-type enzyme and thus, significantly different (t-test; p < 0.01).
Figure 7.
Inhibition of pyruvate carboxylation activity by L-aspartate in the absence of acetyl CoA catalysed by wild-type RePC (■), R427S (●), R427K (○), R472S (▲), and R472K (▽) RePC mutants. Solid lines represent nonlinear regression fits of the data to equation (3).
Table 8.
Inhibition of pyruvate carboxylation activity by L-aspartate
| Enzyme | Apparent Ki (mM) | % Residual activity at saturating aspartate |
|---|---|---|
| WT | 5.0 ± 0.6 | 16 ± 3 |
| R427S | 7 ± 1 | 16 ± 4 |
| R427K | 4.5 ± 0.6 | 36 ± 2 |
| R472S | 7 ± 1 | 12 ± 5 |
| R472K | 7 ± 1 | 21 ± 4 |
Assay conditions: 100 mM Tris-HCl (pH7.8), 30°C, 20 mM NaHCO3, 1 mM MgATP, 5 mM MgCl2, L-aspartate (0 – 60 mM). The reported errors are standard errors of the values of the parameters determined from the nonlinear regression fit of the data in Fig. 7 to equation (3)
DISCUSSION
Analytical ultracentrifugation sedimentation velocity experiments showed that mutation of Arg427 and Arg472 to Ser or Arg427 to Lys had little effect on the stability of the enzymic tetramer. In contrast, destabilisation of the tetrameric structure was observed with the R472K mutant, although it is unclear as to why this fairly conservative mutation would affect the stability of the tetramer. While it is possible that the sub-saturating concentrations of acetyl CoA (100 μM) used in these experiments may not afford complete protection against subunit dissociation due to dilution of the enzyme (3), it would be expected that similar destabilisation of the tetrameric enzyme would be observed with the R472S mutant. Based on the determined Ka for acetyl CoA for the R472S (~1650 μM) and R472K (~2040 μM) RePC mutants, the concentration of acetyl CoA in the ultracentrifugation experiments would result in only 0.1% and 0.07% saturation of the enzymes, respectively, suggesting that this is unlikely to be a factor in the differential stabilisation of the tetramer.
The location of Arg472 in the crystal structure of RePC with ethyl CoA in the allosteric domain is such that this residue does not appear to be involved in any direct intersubunit interactions. In fact, in the presence or absence of ethyl CoA, Arg472 appears to be within interacting distance with only one residue, Glu1027. Since the mutation of Arg472 to Ser would most likely disrupt this interaction whilst it would be retained in the Lys variant, this interaction is presumably contributing little, if any, to the overall stabilisation of the tetramer. One purely speculative possibility that cannot be dismissed is that new residue interactions are formed in the R472K mutant which induces the observed destabilisation.
The proposed importance of Arg427 and Arg472 to the proper binding and orientation of the allosteric activator is supported by the inhibitory effect mutations of these residues have on the activation of the pyruvate carboxylation reaction by acetyl CoA. In the structure of the RePC holoenzyme, Arg427 forms a weak hydrogen bond with the 3′-phosphate group of ethyl CoA (3.3 Å between the δ-N of Arg427 and the oxygen of the 3′-phosphate, Fig. 2b) while Arg472 forms strong hydrogen bonds with the 5′-α-phosphate of ethyl CoA (2.6 Å between the δ-N of Arg472 and one non-bridging oxygens; 2.6 Å between the ω-N of Arg472 and the other non-bridging oxygen, Fig. 8). Compared to the interactions with Arg427, the stronger interactions established between Arg472 and ethyl CoA reasonably explains why the binding of acetyl CoA is more severely affected in the Arg472 RePC mutants resulting in significant increases in the apparent Ka for acetyl CoA.
Figure 8.
Part of the allosteric binding site for acetyl CoA in RePC showing the contacts of Arg427 and Arg472 with the acetyl CoA analogue, ethyl CoA, and adjacent residues.
The Lys mutations of both Arg427 and Arg472 exhibited a more pronounced effect on acetyl CoA binding as compared to the respective Ser mutations. These differential effects most likely arise from the precise positioning of a constellation of residues surrounding acetyl CoA in the allosteric domain. Through the ω-amino group, Arg427 interacts with the carbonyl oxygens of Arg424, Glu425 and Phe426 and Arg472 interacts with the side chain carboxyl group of Glu1027 (Fig 8a). Thus, Arg472 and Arg427 allow for the precise and specific positioning of these residues leading to the specific positioning and orientation of the bound acetyl CoA. When either of the Arg residues are replaced by Ser, the residue-residue and residue-CoA interactions are likely to be lost and the positioning and orientation of the bound acetyl CoA will be determined by any remaining interactions. Replacement of either Arg residue with a Lys could potentially allow for interactions with acetyl CoA, but would not allow for interactions with surrounding residues in the binding pocket. This could lead to the improper positioning of the acetyl CoA in the allosteric site and increased observed Ka values, with the effect being more severe in the case of the R427K mutant.
With the exception of the R427K mutant, the RePC mutants exhibited Hill coefficients for the activation of the pyruvate carboxylation reaction by acetyl CoA similar to that determined for the wild-type enzyme, suggesting that the cooperative mechanism by which acetyl CoA binds to the enzyme still occurs. In the case of R427K however, the Hill coefficient was much lower than that for the wild-type enzyme, suggesting that interactions leading to the cooperative binding of acetyl CoA may have been lost due to the improper positioning of acetyl CoA in the allosteric site. Since mutation of Arg472 does not affect the cooperative binding of acetyl CoA, this suggests that other interactions are important for this facet of activator binding e.g the interactions of acetyl CoA with Asp47 and/or Asn1055 shown in Fig. 2b. The interaction with Asp47 is the most likely candidate since this residue comes from an adjacent subunit in the enzyme tetramer and could thus potentially form key inter-subunit interactions required to transmit the allosteric effects of acetyl CoA binding. Further, mutation of Arg427 increased the catalytic rates of the acetyl CoA-independent carboxylation of pyruvate (k°cat) by 2.2–5.6 fold, whilst mutation of Arg472 resulted in k°cat values that were not significantly different from the wild-type enzyme. This suggests that one or more of interactions between Arg427 and the surrounding residues observed in the absence of the allosteric effector (15) are involved in constraining the structure of the enzyme in a less active conformation.
The effects of mutation of Arg427 and Arg472 on the Km for MgATP and the activation of pyruvate carboxylation by free Mg2+ may have influenced the kcat measurements with acetyl CoA at fixed concentrations of MgATP (1 mM) and free Mg2+ (5 mM). Theoretical values of kcat at saturating concentrations of MgATP and Mg2+ were calculated using the determined values of Km and Ka for R427S, R427K, R472S and R472K, these are 5.2 s−1, 5.2 s−1, 8.5 s−1 and 10.8 s−1 respectively. Comparing these calculated values to the kcat of the wild-type enzyme (10.6 s−1) the decreased values of kcat measured for the R472S and R472K mutants (Table 2) could be possibly be attributed to non-saturating concentrations of MgATP and free Mg2+. On the other hand, the decreased values of kcat measured for the Arg427 mutants were only partly caused by the increases in the Km for MgATP and the Ka for free Mg2+. Even at saturating concentrations of these reaction components, the rate of pyruvate carboxylation for R427S- and R427K-catalysed reactions is 50% of that for wild-type RePC, indicating that the mutation of Arg427 slows a rate-limiting step in the reaction. Pre-steady state studies on pyruvate carboxylases (26, 27) have provided evidence that acetyl CoA enhances the rate of both ATP-cleavage and biotin binding to the BC domain active site. Branson and Attwood (26) concluded that biotin binding was likely to be rate-limiting in the pyruvate carboxylation reaction. Thus, the mutation of Arg427 is likely to affect this step.
Based on the current kinetic analysis, mutations of Arg427 and Arg472 primarily affect reactions in the BC domain. This is confirmed by the marked reduction in the rates of bicarbonate-dependent ATP-cleavage, phosphorylation of ADP by carbamoyl phosphate and biotin carboxylation observed in the mutant catalysed reactions. The effects on the steady-state rates of ATP-cleavage and biotin carboxylation are much greater than on the rates of ADP phosphorylation. It is unlikely that the observed rate reductions in these reactions can be solely attributable to changes in the Km for MgATP and the Ka for free Mg2+ since it has been established that the Km for MgATP in the bicarbonate-dependent ATP cleavage reaction is 5-fold lower than it is for the pyruvate carboxylation reaction and that free Mg2+ has a lower stimulatory effect on ATP-cleavage as the concentration of MgATP approaches saturation (8). In addition, the concentration of free Mg2+ (8 mM) used for determining the rate of ADP phosphorylation will likely minimise any effects attributable to the increased Ka for Mg2+. Whilst biotin is involved in the steady-state ATP-cleavage reaction, it does not directly participate in the ADP phosphorylation reaction but has been clearly shown to enhance the rate of the ADP phosphorylation reaction in PC (24). The larger effects of the mutations on the bicarbonate-dependent ATP-cleavage reaction compared to those on the kcat values for pyruvate carboxylation can also be explained in terms of the effects on biotin binding since Branson and Attwood (26) and Branson et al. (27) noted that the biotin binding step was more rate-limiting in the bicarbonate-dependent ATP-cleavage reaction than in the pyruvate carboxylation reaction.
Arg427 and Arg472 are approximately 40 Å and 47 Å from the ATP-binding site in the BC domain and are most likely not directly influencing the positioning of catalytically important residues in the BC domain active site. In fact, based on the RePC holoenzyme structure (21) the presence or absence of ethyl CoA had no effect on the positions of the catalytically important residues Glu218, Lys 245, Arg301 and Glu305 (7) relative to ATP-γ-S. Therefore, it is probable that the interactions between Arg427 and Arg472, acetyl CoA and nearby residues in the allosteric domain promote the binding of biotin at the BC domain active site. In the first structure of RePC to be determined (14) the asymmetric enzymic tetramer contained only one pair of subunits on one face of the tetramer optimally configured for inter-subunit catalysis. Only this pair of subunits had ethyl CoA bound at the allosteric sites. While it was initially assumed that the binding of acetyl CoA to these subunits induced the asymmetrical arrangement of the tetramer, Lietzan et al. (15) showed that RePC crystalized as an asymmetrical tetramer in the absence of an allosteric activator. These differences in the positions of amino acid residues in subunits that have an activator bound at the allosteric site compared to subunits that do not are difficult to interpret as it is not clear whether the inherent asymmetry of the tetramer has caused the changes or the binding of the allosteric activator. Unfortunately, parts of the structure of RePC determined in the absence of allosteric activator are poorly defined, including the polypeptide region containing Arg427 and Arg472. This makes a comparison of the positions and interactions of Arg427 and Arg472 between this structure and that with an allosteric activator bound impossible. To fully understand the roles of Arg427 and Arg472 in the allosteric action of acetyl CoA it will be necessary to produce a well-resolved structure of RePC determined in the absence of allosteric activators.
The lack of effect the mutations of Arg427 and Arg472 on the Ki for L-aspartate indicates that these residues are most likely not involved in the binding of L-aspartate. In fact, the only observed effect on L-aspartate inhibition was seen with the R427K mutant, which is proposed to promote the adoption of a more catalytically active state of the tetrameric enzyme thereby leading to enhanced acetyl CoA-independent activity and reduced cooperativity. For this reason, L-aspartate may have less inhibitory effect on this mutant, even at saturating concentrations.
In this work we have shown that both Arg427 and Arg472 are important for the binding of the allosteric activator and, as predicted from the structure of RePC, Arg472 plays the most important role in this process. Unexpectedly, we found that the more conservative mutation of the Arg to Lys at both positions resulted in greater detrimental effects on acetyl CoA binding than less conservative mutations to Ser. We have proposed that whilst the Lys mutants are still capable of interacting with acetyl CoA, the positioning of the Lys residues via interactions with proximal amino acid residues are lost, leading to the improper positioning of acetyl CoA. In the Ser mutants, the direct interaction with acetyl CoA is lost, leaving the positioning of the bound acetyl CoA to be determined by secondary interactions with other amino acid residues. The effects of mutation of Arg427 and Arg472 are not restricted solely to acetyl CoA binding. Major effects on reactions occurring in the BC domain active site suggest that Mg2+ binding, ATP-cleavage and biotin carboxylation are also affected by mutations incorporated in the allosteric domain. We propose that the mutations produce these effects in the BC domain by interfering with the binding of biotin in this active site. Arg427 and Arg472 do not appear to be directly in the binding of the allosteric inhibitor L-aspartate.
The primers used to generate the mutant forms of RePC (R427S, R427K, R472S and R472K) are shown in Table S1 of Supplemental Information. Supplemental materials may be accessed free of charge online at http://pubs.acs.org.
Supplementary Material
Table 5.
Effects of acetyl CoA on the rates of the bicarbonate-dependent MgATP cleavage reaction
| Enzyme | kcat (s−1 ) (saturating acetyl CoA) | % | kcat (s−1) (no acetyl CoA) | % |
|---|---|---|---|---|
| WT | 1.6 ± 0.1 | 100 | 0.19 ± 0.02 | 100 |
| R427S | 0.017 ± 0.001 | 1.1 | 0.011 ± 0.001 | 7.5 |
| R427K | 0.0064 ± 0.0002 | 0.4 | 0.0016 ± 0.0003 | 0.8 |
| R472S | 0.0090 ± 0.0005 | 0.6 | 0.0023 ± 0.0007 | 1.2 |
| R472K | 0.009 ± 0.002 | 0.6 | 0.0017 ± 0.0005 | 0.9 |
Assay conditions: 100 mM Tris-HCl (pH7.8), 30°C, 20 mM NaHCO3, 6 mM MgCl2, 1 mM ATP, in the absence or presence of saturating concentrations of acetyl CoA (see Materials and Methods). The reported errors are standard deviation of the means of three separate determinations of the apparent kcat values.
Acknowledgments
Funding: This work was supported by the National Institute of Health grant GM070455 to W.W.C., M.St.M., J.C.W., and P.V.A. and an NIH award F32DK083898 from the National Institute of Diabetes and Digestive and Kidney Diseases to T.N.Z.
Abbreviations
- acetyl CoA
acetyl coenzyme A
- PC
pyruvate carboxylase
- RePC
Rhizobium etli pyruvate carboxylase
- BC
biotin carboxylase
- CT
carboxyl transferase
- BCCP
biotin carboxyl carrier protein
Contributor Information
Abdussalam Adina-Zada, Email: abdul.adina-zada@uwa.edu.au.
Chutima Sereeruk, Email: sereeluck@hotmail.com.
Sarawut Jitrapakdee, Email: scsji@mucc.mahidol.ac.th.
Tonya N. Zeczycki, Email: zeczyckit@ecu.edu.
Martin St. Maurice, Email: martin.stmaurice@marquette.edu.
W. Wallace Cleland, Email: cleland@enzyme.wisc.edu.
John C. Wallace, Email: john.wallace@adelaide.edu.au.
Paul V. Attwood, Email: pattwood@cyllene.uwa.edu.au.
References
- 1.Jitrapakdee S, Wallace JC. Structure, function and regulation of pyruvate carboxylase. Biochem J. 1999;340:1–16. doi: 10.1042/bj3400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace J. Distribution and biological functions of pyruvate carboxylase in nature. In: Keech D, Wallace J, editors. Pyruvate Carboxylase. CRC Press; Boca Raton: 1985. pp. 5–64. [Google Scholar]
- 3.Adina-Zada A, Zeczycki TN, Attwood PV. Regulation of the structure and activity of pyruvate carboxylase by acetyl CoA. Arch Biochem Biophys. 2012;519:118–130. doi: 10.1016/j.abb.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn MF, Encarnación S, Araíza G, Vargas MC, Dávalos A, Peralta H, Mora Y, Mora J. Pyruvate carboxylase from Rhizobium etli: mutant characterization, nucleotide sequence, and physiological role. J Bacteriol. 1996;178:5960–5970. doi: 10.1128/jb.178.20.5960-5970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attwood PV, Wallace JC. Chemical and catalytic mechanisms of carboxyl transfer reactions in biotin-dependent enzymes. Acc Chem Res. 2002;35:113–120. doi: 10.1021/ar000049+. [DOI] [PubMed] [Google Scholar]
- 6.Jitrapakdee S, StMaurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeczycki TN, Menefee AL, Adina-Zada A, Jitrapakdee S, Surinya KH, Wallace JC, Attwood PV, Maurice M, Cleland WW. Novel insights into the biotin carboxylase domain reactions of pyruvate carboxylase from Rhizobium etli. Biochemistry. 2011;50:9724–9737. doi: 10.1021/bi2012788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attwood PV, Graneri BD. Bicarbonate-dependent ATP cleavage catalysed by pyruvate carboxylase in the absence of pyruvate. Biochem J. 1992;287:1011–1017. doi: 10.1042/bj2871011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legge GB, Branson JP, Attwood PV. Effects of acetyl CoA on the pre-steady-state kinetics of the biotin carboxylation reaction of pyruvate carboxylase. Biochemistry. 1996;35:3849–3856. doi: 10.1021/bi952797q. [DOI] [PubMed] [Google Scholar]
- 10.Zeczycki TN, Menefee AL, Jitrapakdee S, Wallace JC, Attwood PV, Maurice M, Cleland WW. Activation and inhibition of reactions occurring in the biotin carboxylase domain of pyruvate carboxylase from Rhizobium etli. Biochemistry. 2011;50:9694–9707. doi: 10.1021/bi201276r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Attwood PV, Wallace JC. The carboxybiotin complex of chicken liver pyruvate carboxylase a kinetic analysis of the effects of acetyl CoA, Mg2+ ions and temperature on its stability and on its reaction with 2-oxobutyrate. Biochem J. 1986;235:259–264. doi: 10.1042/bj2350359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adina-Zada A, Jitrapakdee S, Surinya KH, McIldowie MJ, Piggott MJ, Cleland WW, Wallace JC, Attwood PV. Insights into the mechanism and regulation of pyruvate carboxylase by characterisation of a biotin-deficient mutant of the Bacillus thermodenitrificans enzyme. Int J Biochem Cell Biol. 2008;40:1743–1752. doi: 10.1016/j.biocel.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Jitrapakdee S, Adina-Zada A, Besant PG, Surinya KH, Cleland WW, Wallace JC, Attwood PV. Differential regulation of the yeast isozymes of pyruvate carboxylase and the locus of action of acetyl CoA. Int J Biochem Cell Biol. 2007;39:1211–1223. doi: 10.1016/j.biocel.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.St Maurice M, Reinhardt L, Surinya KH, Attwood PV, Wallace JC, Cleland WW, Rayment I. Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science. 2007;317:1076–1079. doi: 10.1126/science.1144504. [DOI] [PubMed] [Google Scholar]
- 15.Lietzan AD, Menefee AL, Zeczycki TN, Kumar S, Attwood PV, Wallace JC, Cleland WW, St Maurice M. Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli. Biochemistry. 2011;50:9708–9708. doi: 10.1021/bi201277j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang S, Tong L. Crystal structures of human and S. aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat Struct Mol Biol. 2008;15:295–302. doi: 10.1038/nsmb.1393. [DOI] [PubMed] [Google Scholar]
- 17.Yu LP, Xiang S, Lasso G, Gil D, Valle M, Tong L. A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure. 2009;17:823–832. doi: 10.1016/j.str.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasso G, Yu LP, Gil D, Xiang S, Tong L, Valle M. Cryo-EM analysis reveals new insights into the mechanism of action of pyruvate carboxylase. Structure. 2010;18:1300–1310. doi: 10.1016/j.str.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeczycki TN, St Maurice M, Jitrapakdee S, Wallace JC, Attwood PV, Cleland WW. Insight into the carboxyl transferase domain mechanism of pyruvate carboxylase from Rhizobium etli. Biochemistry. 2009;48:4305–4313. doi: 10.1021/bi9003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman-Smith A, Turner DL, Cronan JE, Jr, Morris TW, Wallace JC. Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. Biochem J. 1994;302:881–887. doi: 10.1042/bj3020881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rylatt DB, Keech DB, Wallace JC. Pyruvate carboxylase: isolation of the biotin-containing tryptic peptide and the determination of its primary sequency. Arch Biochem Biophys. 1977;183:113–122. doi: 10.1016/0003-9861(77)90425-8. [DOI] [PubMed] [Google Scholar]
- 22.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attwood PV, Cleland WW. Decarboxylation of oxalacetate by pyruvate carboxylase. Biochemistry. 1986;25:8191–8196. doi: 10.1021/bi00373a011. [DOI] [PubMed] [Google Scholar]
- 24.Attwood PV, Graneri BD. Pyruvate carboxylase catalysis of phosphate transfer between carbamoyl phosphate and ADP. Biochem J. 1991;273:443–448. doi: 10.1042/bj2730443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adina-Zada A, Hazra R, Jitrapakdee S, Zeczycki TN, StMaurice M, Cleland WW, Wallace JC, Attwood PV. Probing the allosteric activation of pyruvate carboxylase using 2′,3′-O-(2,4,6-Trinitrophenyl) adenosine 5′-triphosphate as a fluorescent mimic of the allosteric activator acetyl CoA. Arch Biochem Biophys. 2011;509:117–126. doi: 10.1016/j.abb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branson JP, Attwood PV. Effects of Mg2+ on the pre-steady-state kinetics of the biotin-carboxylation reaction of pyruvate carboxylase. Biochemistry. 2000;39:7480–7491. doi: 10.1021/bi992825v. [DOI] [PubMed] [Google Scholar]
- 27.Branson JP, Nezic M, Wallace JC, Attwood PV. Kinetic characterisation of yeast pyruvate carboxylase iosozyme Pyc1. Biochemistry. 2002;41:4459–4466. doi: 10.1021/bi011888m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







