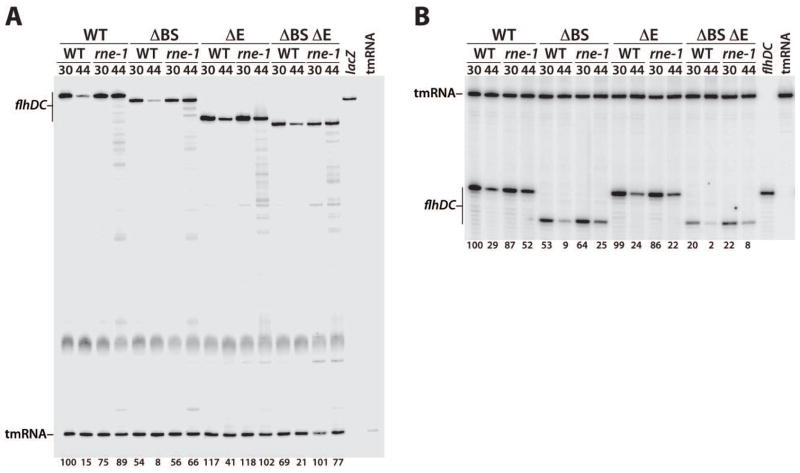
Fig. 9. Quantitative primer extension analysis of WT, ΔBS, ΔE and ΔBS ΔE transcript decay.
RNA was isolated from WT and rne mutant strains containing chromosomally integrated flhD'-'lacZ fusions (A) or truncated flhD-tR2 constructs (B). Cells were grown at 30°C and harvested before or after a shift to the non-permissive temperature (44°C).
A. Primers specific for lacZ mRNA and tmRNA were annealed simultaneously to total cellular RNA and extended by reverse transcriptase. Levels of full-length flhDC transcripts were normalized to tmRNA levels. The level of RNA for the WT transcript in the WT (rne+) genetic background at 30°C was set to 100%. Normalized RNA levels are shown at the bottom of each lane. Control reactions with only the lacZ or tmRNA primers are marked.
B. Primers specific for flhDC leader and tmRNA were annealed simultaneously to total cellular RNA and extended by reverse transcriptase. Levels of full-length flhDC transcripts were normalized to tmRNA levels. The level of RNA for the WT transcript in the WT (rne+) genetic background at 30°C was set to 100%. Normalized RNA levels are shown at the bottom of each lane. Control reactions with only the flhDC leader or tmRNA primers are marked. Since the flhDC leader primer hybridizes upstream of the ΔE deletion, the length of the primer extension products for WT and ΔE, as well as for ΔBS and ΔBS ΔE, are the same.