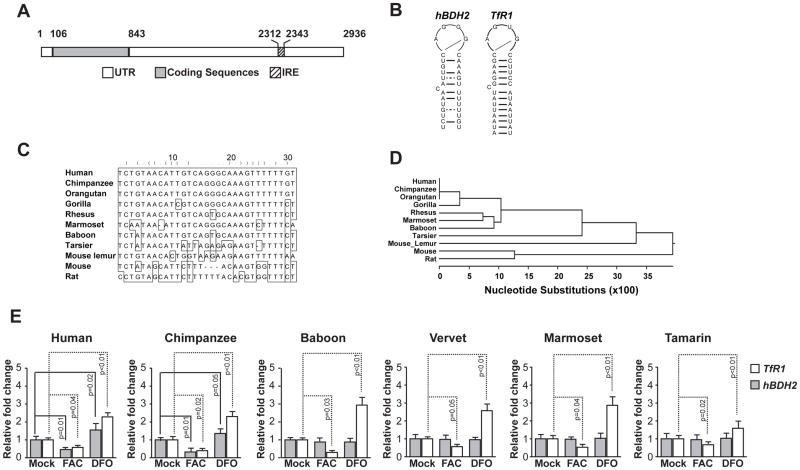
Figure 3. Structure, sequence, and phylogenic conservation of hBDH2 3′-UTR IRE.
(A) Location of the hBDH2 IRE in the 3′-UTR. IRE. Black boxes indicate exons and the hatched box indicates the location of the IRE. (B) Predicated secondary structures of 3′-UTR IREs of hBDH2 and hTfR1. (C) Multispecies alignment of hBDH2 3′-UTR IRE sequences. (D) Phylogenetic distribution of bdh2 IRE. (E) Iron-dependent regulation of hBDH2 mRNA is restricted to members of the hominidae family. TfR1 and hBDH2 mRNAs were quantified by real time PCR in transformed human and primate liver cell lines 16 hours after treatment with FAC or DFO. The value on y-axis was set at 1.0 for the mRNA levels in untreated cells. The relative mRNA levels in each sample were normalized to eif2b2 (an internal standard for RT-PCR analysis of primate mRNA samples). Positive control includes TfR1, whose expression is affected by iron. Results shown are the average means of three independent experiments with error bars depicting SD.