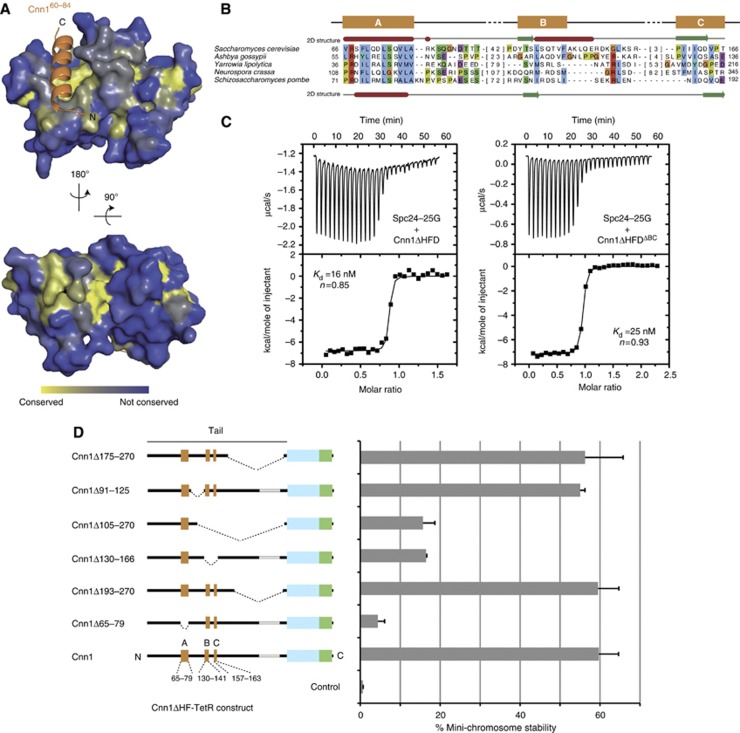
Figure 5.
Functional dissection of the Cnn1 N-tail. (A) Conservation of Spc24-25 plotted onto the structure of the Spc24-25 globular domain. Highly conserved surfaces are coloured in yellow and less conserved surfaces in blue. Note the presence of conserved residues in the Cnn1 binding pocket and on the opposite site of the globular domain. (B) Multiple sequence alignment of an N-terminal region in Cnn1 homologues. S. cerevisiae (above) and S. pombe (below) secondary structure predictions are derived from the Jpred 3 server (α helices in red and β strands in green) (Cole et al, 2008). Background colouring of the residues is based on the Clustalx colouring scheme. The number of omitted residues is indicated in brackets. See Supplementary Figure S5 for alignment of more sequences. (C) Isothermal titration calorimetry performed by titrating the Spc24-25 globular domain (Spc24-25G) with Cnn1 deleted of the histone-fold domain (Cnn1ΔHFD) on the left, or additionally deleted of residues 130–166 (Cnn1ΔHFDΔBC) on the right. Note the very similar Kd values, estimated by fitting a non-linear curve to the derived binding isotherms. (D) Stability of acentric URA3 mini-chromosomes segregated through artificial recruitment of Cnn1ΔHF-TetR. The respective Cnn1 construct is indicated in the left panel (dotted lines indicate the position of the deletion), the corresponding mini-chromosome stability on the right. Error bars denote s.e.m. (n=3).