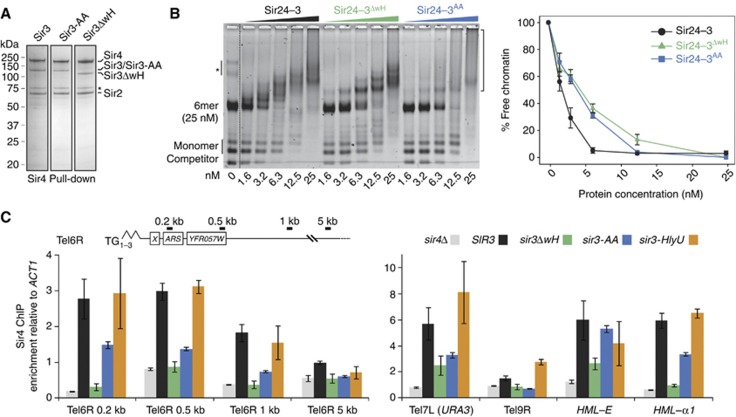
Figure 5.
Sir3 wH homodimerization is critical for loading or stable binding of Sir complex onto chromatin in vivo. (A) Hi5 insect cells were co-infected with Sir2, Sir4 and the indicated Sir3 expressing virus. Sir4 was affinity purified as described previously (Martino et al, 2009) and a sample elution was loaded on an SDS–PAGE and stained with Coomassie blue. The asterisk indicates a common Sir4 degradation product. (B) The holo-Sir complex or mutant complexes containing either Sir3ΔwH (aa 1–850) or Sir3-AA (L861A and V909A) were titrated into a constant amount of 6-mer of nucleosomes. The images are representative of at least three independent experiments; quantifications show the mean value ±s.e.m. of the % of unbound chromatin compared to the input. The asterisk indicates a contaminant DNA, and the bracket on the right indicates Sir/chromatin complexes. (C) Sir4 ChIP analysis of the same strains used for silencing assay in Figure 3E, the amount of precipitated DNA was measured by qPCR. Enrichment over the ACT1 locus was normalized to the input.