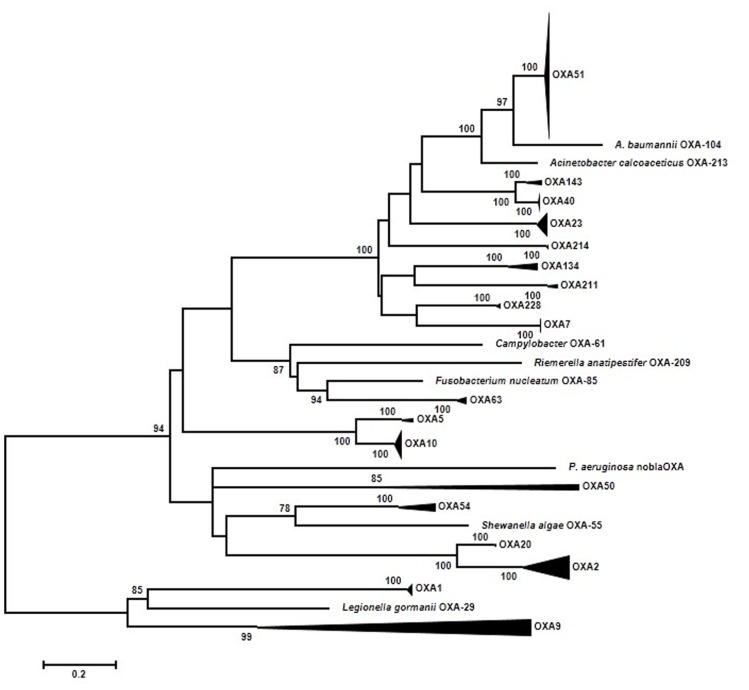
FIGURE 2.
Summarized representation of the maximum-likelihood tree of OXA genes obtained with the K2P + GI model of evolution. Multiple nucleotide sequence alignments were obtained using the corresponding amino acid sequences with Muscle (Edgar, 2004a,b). Maximum-likelihood trees were obtained using the most appropriate model of evolution resulting from the comparison of Bayesian Information Criterion (BIC) values for 24 alternative models including gamma-distributed heterogeneous rates of evolution among and invariant sites. Support of the inferred clusters was evaluated with 1000 bootstrap replicates; BS values >70% are indicated. All these methods were used as implemented in MEGA 5 (Tamura et al., 2011). The scale bar corresponds to substitutions/site. The role of natural selection was investigated at the codon level by analyzing the difference between dN–dS substitution rates at different positions. Based on the corresponding maximum likelihood trees using a Muse–Gaut model (Muse and Gaut, 1994) of codon substitution and the Tamura–Nei model of nucleotide substitution (Tamura and Nei, 1993). Computation of dN/dS was performed with the Hyphy software package (Kosakovsky Pond et al., 2005). Additionally, two tests of neutrality were applied to the three data sets: the Fisher’s exact test of neutrality and a test based on bootstrapping (1000 replicates) of dN–dS values from which a variance of the corresponding statistic is derived in order to test the null hypothesis of neutrality (dN = dS). These tests were performed using MEGA 5 (Tamura et al., 2011).