Fig. 4.
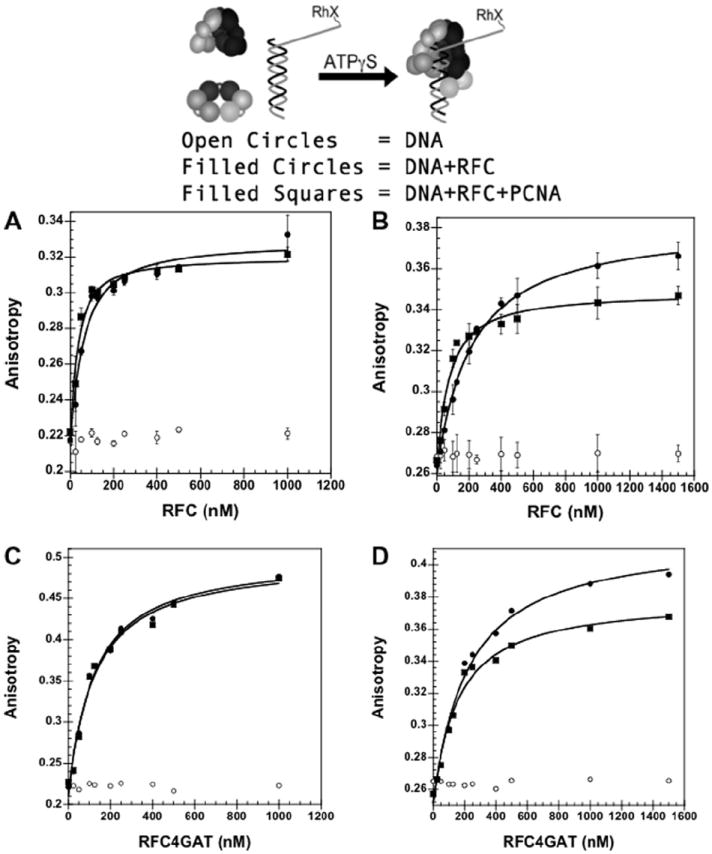
Equilibrium DNA binding assay in the presence and absence of 750 mM maltose. DNA-RhX fluorescence anisotropy is plotted. The anisotropy increases upon binding by RFC or RFC▪PCNA. (A) wt RFC▪DNA binding in the absence of maltose. (B) wt RFC▪DNA binding in the presence of maltose. (C) RFC4GAT▪DNA binding in the absence of maltose. (D) RFC4GAT▪DNA binding in the presence of maltose. The anisotropy of RhX at 605 nm is plotted as a function of RFC concentration for free DNA (open circles), RFC + DNA (filled circles) and RFC + PCNA + DNA (squares). Data were fit to Eq. (4) (Materials and methods) using Kaleidagraph [14,17] to obtain Kd values and are the average of three independent experiments for (B) and (C). Data in (D) and (E) are single experiments. DNA-RhX, RFC, and PCNA were added sequentially to a solution of assay buffer and ATPγS. Final reaction conditions: 30 mM HEPES pH 7.5, 150 mM NaCl, 2 mM DTT, 8 mM MgCl2, 0.5% glycerol, 0.5 mM ATPγS, 1 μM PCNA (without maltose) or 1.5 μM PCNA (with maltose), and 0–1 μM RFC (without maltose) or 0–1.5 μM RFC (with maltose).
