Abstract
Pluripotent stem cells can be created successfully through the inner cell mass (ICM), nuclear transfer, and defined-factor induction. Unfortunately, the epigenetic characteristics of the cells produced are poorly understood. In this article, we compared expression levels of enzymes involved in epigenetic modifications across six pluripotent stem cell lines. Six of the 11 genes evaluated here (Dnmt3a, Dnmt3b, Tet1, Ezh2, Mll1, and Lsd1) showed abnormally low levels of expression in the two germ-line chimeric induced pluripotent stem cell (iPSC) lines. We also conducted locus-specific analysis of DNA methylation at 9 loci. Although iPSCs did express Oct4, the Oct4 promoter region was shown to have a higher level of DNA methylation. The Xist and Line-1 repeating sequences differed relatively little in methylation level across the cell lines, but Peg3, Peg10, and H19 exhibited high degrees of variation in the pattern of DNA methylation. Meg3 in the Dlk1–Dio3 imprinting cluster was incompletely methylated in embryonic stem cells (ESCs) and nuclear transfer (nt) ESCs. However, in germ-line chimeric iPSCs, Meg3 was almost entirely methylated. ESC and ntESC lines showed twice as much Meg3 expression than in the iPSC lines. The genomic 5mC contents detected by reverse-phase high-performance liquid chromatography (HPLC) indicated that, despite their germ-line chimeric abilities, iPSCs remained incompletely reprogrammed, even though no direct evidence is shown here.
Introduction
Pluripotent stem cells can differentiate into any type of cell or tissue in the body. They have a myriad of potential applications in organ regeneration and repair and in the treatment of diseases (Chien, 2008). Years ago, these cells could be obtained only from human and mouse embryos, which raised ethical concerns in many circles (Evans and Kaufman, 1981; Thomson et al., 1998). In recent years, however, biologists have derived controversy-free stem cells via somatic cell reprogramming (Hochedlinger and Jaenisch, 2006). There are two types of reprogrammed somatic cells that can self-renew and generate any type of cell in the body in vivo and in culture: (1) nuclear transfer embryonic stem cells (ntESCs) and defined-factor induced pluripotent stem cells (iPSCs) (Hochedlinger and Jaenisch, 2003). Nuclear transfer technology can be used to reprogram somatic cells after injection into an enucleated oocyte. This can be used to derive patient-specific pluripotent cells, and it is called therapeutic cloning (Hochedlinger and Jaenisch, 2003). Mouse iPSCs can be generated by co-expression of three or four transcription factors directly from fibroblasts. They hold great promise in research and cell therapy (Takahashi et al., 2007; Robinton and Daley, 2012). Previous research has shown that both ntESCs and iPSCs are transcriptionally and functionally equivalent to normal ESCs. However, several recent studies that compared iPSCs to normal ESCs with respect to gene transcription, copy number variation, DNA methylation, and chromatin modification showed iPSCs reprogrammed from somatic cells to have abnormal demethylation and retain residual the genomic imprinting status of the donor cells (Chin et al., 2009; De Carvalho et al., 2010; Polo et al., 2010; Gaspar-Maia et al., 2011; Hussein et al. 2011; Lister et al. 2011; Nishino et al. 2011).
Recent studies have evaluated the quality of pluripotent stem cells (Kim et al., 2010). Some iPSCs retain epigenetic imprints of their parental cell types. The selection of certain donor cell types can used to facilitate erasure of methylation status and avoid aberrant methylation reprogramming (Kim et al., 2011). These studies showed ntESCs and iPSCs to have epigenetic states similar to that of normal ESCs rather than those of somatic cells. The activation of the imprinted Dlk1–Dio3 region has been shown to be positively correlated with the level of pluripotency of iPSCs generated by tetraploid complementation. Researchers remain unsure about the extent to which the three pluripotent stem cell lines (ESCs, ntESCs, and iPSCs) differ from each other with respect to genomic imprinting at specific DNA methylation loci.
To evaluate the differences in epigenetic modification among the three pluripotent stem cell types and determine the epigenetic characteristics of germ-line chimeric iPSCs, we quantified the transcription levels of several enzymes involved in epigenetic modification using real-time PCR with a focus on differences of DNA methylation (5mC) at the genomic level. We evaluated the methylation status of maternally imprinted loci (Peg3, Peg10) and two paternally imprinted loci (H19 and IGF2R) using bisulfite sequencing. The iPSCs showed incomplete reprogramming in this respect.
Materials and Methods
Culture of ESCs or iPSCs
Blastocyst-derived ESCs (IVPE1, IVPE2), ntESCs (ntES1, ntES2) derived from cloned embryos, and germ-line chimeric iPSCs lines (IP2-19, IP2-33) were purchased from Beijing Stem Cell Bank, P.R. China. These cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 15% ESC-qualified fetal bovine serum (FBS), 1×nonessential amino acids (NEAA), 1×l-glutamine, 5.5 mM 2-mercaptoethanol, and 1,000 units/mL of leukemia inhibitory factor (LIF) (Millipore). All of the materials used for ESC culture and passage were purchased from Gibco or Invitrogen.
Eight-cell blastocyst injection
iPSCs were digested and dispersed into single cells using trypsin. iPSCs with green fluorescence were picked out under a Olympus X71 (U-RFL-T) fluorescence microscope. A hypodermic needle was then used to transfer these cells into zonae pellucidae of the eight-cell stage blastocysts, ensuring direct contact between the iPSCs and the embryonic blastomeres. The embryos were cultivated in vitro for 48 h and then observed under the Olympus X71 (U-RFL-T) microscope.
RNA extraction and quantitative real-time RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen). Two micrograms of RNA was reverse-transcribed using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega) and dT18 primer according to the manufacturer's instructions. Real-time quantitative PCR (qPCR) reactions were set up in triplicate using Brilliant II SYBR Green QPCR Master Mix (Toyobo) and run on an Mx3000P QPCR System (Stratagene). Table S1 summarizes primer sequences and amplification conditions. (Supplementary Data are available at www.liebertpub.com/cell/.)
High-performance liquid chromatography
For determination of genomic 5 mC content, DNA solution (10 μg) was mixed with hydrofluoric acid at a 1:9 ratio by volume. The mixture was hydrolyzed at 85°C for 24 h, dried completely using a Concentrator Plus (Eppendorf AG), dissolved in the mixed mobile phase (PICB7, 6.8 mM, pH 4) of high-performance liquid chromatography (HPLC), and then filtered through a 0.22-μm filtration membrane. The sample was divided into three 20-μL subsamples, each of which was passed through a HPLC column (150×2.1 mm, 5 μm, Hypersil BDS C18 Thermo) at a flow rate of 0.3 mL/min. A built-in A280 UV detector (Agilent 1100) was used to determine the characteristic absorption curve. Each subsample was tested three times. SPSS 19.0 was used to calculate 5mC content using to the area of the absorption curve 5mC/(5mC+C).
Determination of genomic 5mC content
Bisulfite genomic DNA was extracted using the procedures previously described. One microgram of DNA solution was treated with an EZ DNA Methylation-Gold Kit (Zymo Research) according to the manufacturer's Instructions. Amplified products were purified using a gel extraction kit, then cloned into the PMD19-T vector (Takara) and sequenced with M13 primers. Table S2 summarizes primer sequences and amplification conditions.
Karyotype analysis
Karyotype analysis was carried out using standard mouse chromosome analysis protocols.
Results
Confirmation of the pluripotent state of ntESCs and iPSCs
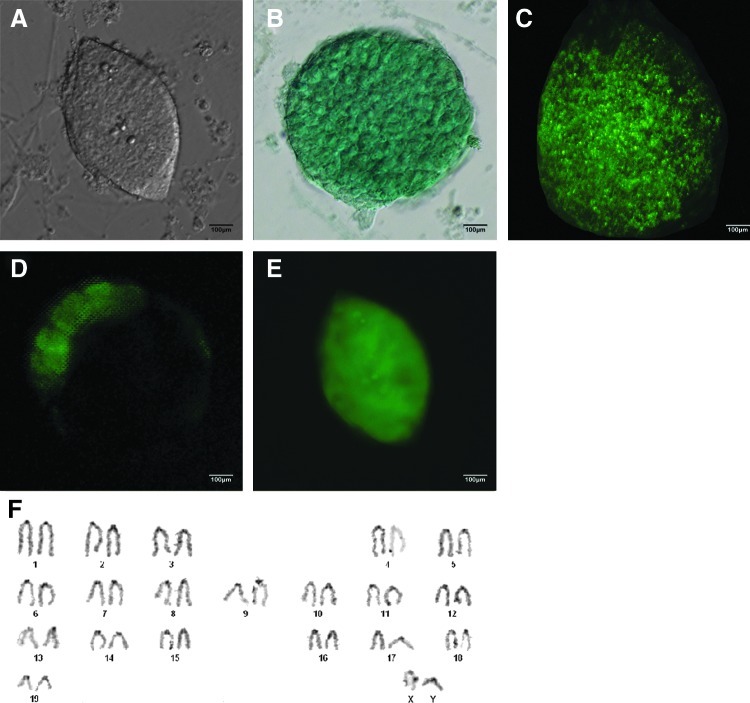
The purchased ESC, ntESC, and iPSC lines were cultured for five or six generations before being used in experiments. Six pluripotent stem cell lines were cultured in ESC standard medium with the CF1 mouse-derived feeder for at least 25 passages. The two germ-line chimeric iPSC lines went through 30 passages and retained normal ESC-like morphology with a high nucleus-to-cytoplasm ratio and prominent nucleoli (Fig. 1A) (Zhao et al. 2009). All six lines of pluripotent cells expressed the cell-surface markers alkaline phosphatase (Fig. 1B), and stage-specific embryonic antigen-1 (SSEA-1) (Fig. 1C), exactly like standard mouse ESCs do. Both of the iPSC lines also expressed the Oct4 green fluorescent protein (GFP) (Fig. 1E). We made further observations on iPSC chimeric blastocysts. Individual cell lines were injected into blastocysts at the eight-cell stage, yielding high chimeric rates (86.0±8.4%) (n=10) in the inner cell mass (ICM) of the blastocyst (Fig. 1D). Two iPSC lines exhibited the normal XY karyotype (Fig. 1F). We also examined the mRNA expression of pluripotency markers Oct4 and Nanog as well (Fig. 2). These experiments suggest that these pluripotent cell lines retained their pluripotency despite five to six extra in vitro passages postpurchase.
FIG. 1.
Characterization of ESCs, ntESCs, and iPSCs. The purchased ES, ntES, and iPS cell lines were cultured for five or six generations before being used for experiments. The six pluripotent cell lines underwent 25–30 generations. Cell morphology resembled that of the ESCs. The expression of pluripotency markers was observed. (A) ESC-like cloned cells. (B) Represententive AP staining of iPSCs. (C) SSEA-1. (D) Eight-cell blastocyst injection of iPSCs. The iPSCs were delivered into embryos at the eight-cell stage and cultured in vitro for 48 h. The chimeric rate of the ICM blastocyst was 86.0±8.4% (n=10), where chimeric rate=ICM fluorescence area/ICM total area (the analysis was carried out with OLYMPUS cellSens Standard Vers1.5). (E) iPSCs were GFP positive, indicating successful expression of Oct4-GFP. (F) Karyotype analysis. The iPS2-19 and iPS2-33 cells at passage 30 showed a normal 40 XY karyotype.
FIG. 2.
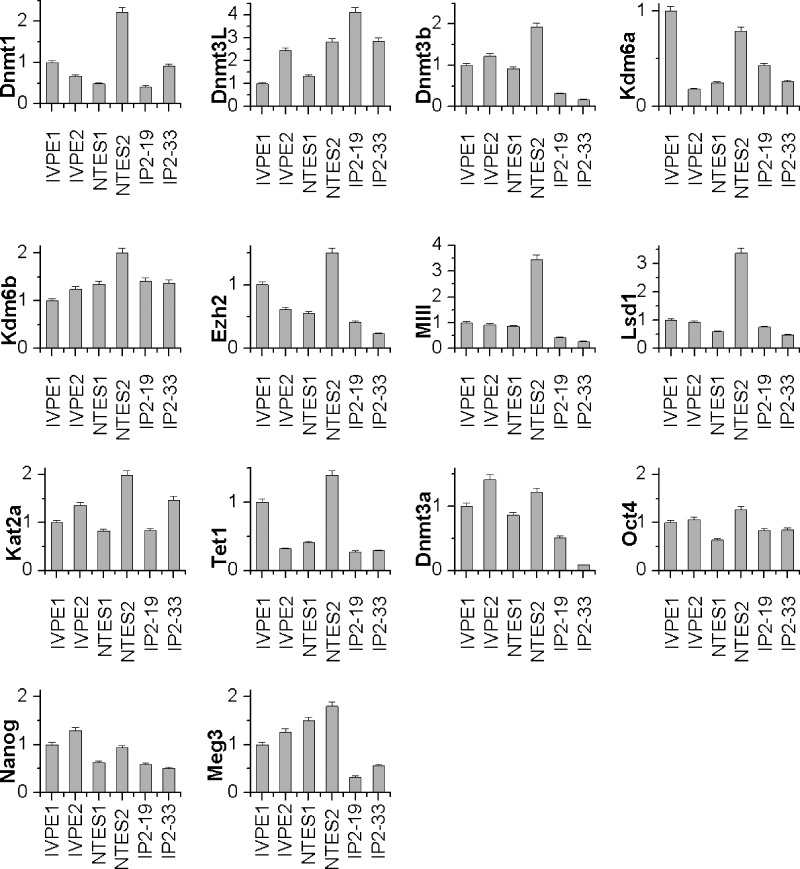
Real-time PCR detection of enzymes pertaining to epigenetic modifications. All samples were normalized against the internal control (β-actin), gene expression value of sample IVPE1 was set to 1, and the other cell lines were compared to sample IVPE1 to produce quantitative values. Each sample was analyzed three times.
Quantification of genes involved in epigenetic modification
Real-time PCR detection of enzymes involved with epigenetic modifications was performed. To compare epigenetic differences among the six pluripotent cell lines, we assigned numeric values to the enzymes involved with epigenetic modifications. Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l are involved with DNA methylation (Okano et al., 1999). Tet1 was involved in DNA demethylation (Tahiliani et al., 2009). Ezh2, Kdm6b, and Kdm6a are associated with H3K27me3 (Greer and Shi, 2012). mll1 and LSDI are connected to H3K4me (Whyte et al., 2012). Kat2a is associated with histone Ac (Smith and Shilatifard, 2010). Samples were normalized against the internal control (β-actin), and the gene expression value of sample IVPE1 was set to 1. The results of this analysis are shown in Figure 2. As indicated by the concentration of relevant mRNA sequences, all enzymes exhibited differences in expression across the different cell lines. ntES2 cells showed higher levels of expression of the examined epigenetic modification enzymes. For example, LSD1 encodes a flavin-dependent monoamine oxidase, which can demethylate mono- and dimethylated lysines, specifically histone 3 and lysines 4 and 9 (H3K4 and H3K9) (Adamo et al., 2011). Mll1 encodes a multiprotein complex that mediates both methylation of Lys-4, which is part of the histone H3 (H3K4me) complex. It also mediates the acetylation of Lys-16, which is part of histone H4 (H4K16ac) (Tahiliani et al., 2009). The ntES2 cell line had LSD1 and MII1 expression levels at least three times higher than the other five cell lines. However, some genes showed similar expression levels across all six cell lines. For example, Kdm6b, a demethylation enzyme of histone H3 K27me3 modification, showed the least amount of intercell line variations in the level of expression (Kooistra and Helin, 2012). Kat2a, associated with histone Ac, also appeared to be more or less equally expressed in all six cell lines.
Genomic cytosine methylation content detected by reverse-phase HPLC
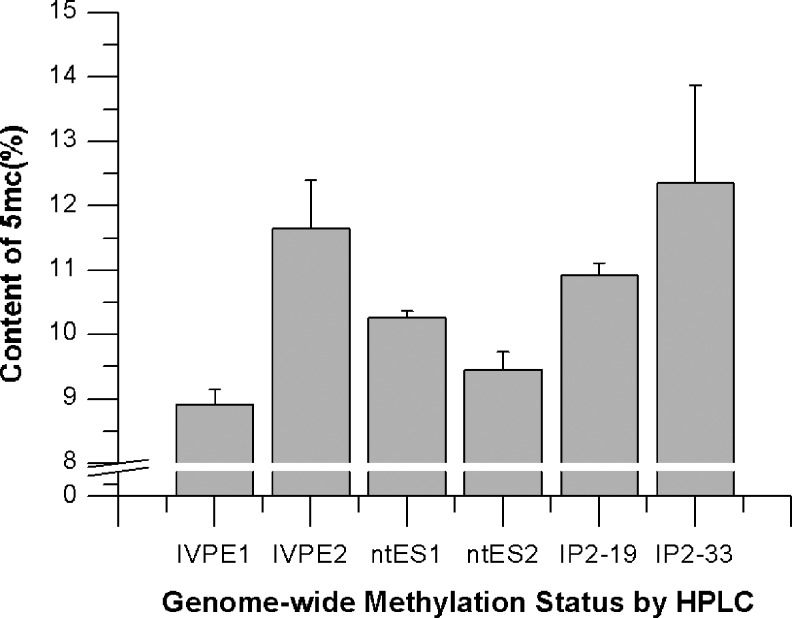
We examined differences in DNA methylation across six pluripotent cell lines. Extracted DNA molecules were dissolved in hydrofluoric acid and then passed through reverse-phase HPLC columns to determine genomic 5mC concentrations (Tahiliani et al., 2009). The results are shown in Figure 3. The degree of DNA methylation differed significantly between the two IVP cell lines (8.9% and 11.7%). The ntESC lines showed similar levels of methylation (10.3% and 9.5%). The iPSC lines showed the highest levels of methylation (10.9% and 12.4%).
FIG. 3.
Genomic cytosine methylation content detected by reverse-phase HPLC: Genomic 5mC content of the six pluripotent cell lines. The 5mC content was calculated according to the area of the absorption curve 5mC/(5mC+C). Each sample was analyzed three times.
FIG. 4.
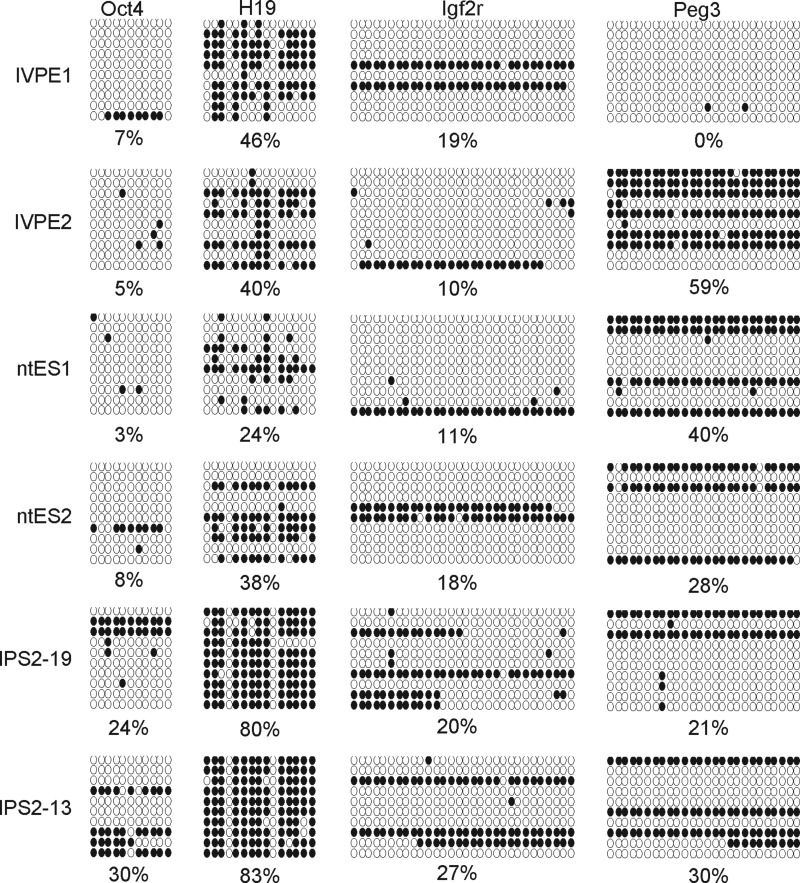
Bisulfite analysis of locus-specific DNA methylation. Genes detected include Oct4, H19, Igf2r, and Peg3. Methylation profiles of their CpG dinucleotides were generated. Scores for the methylation of each CpG were obtained by sequencing PCR clones derived from bisulfite-treated genomic DNAs. (Filled circles) Methylated CpGs; (open circles) unmethylated CpGs. Each row in each cell line represents each sequenced clone of each cell line.
FIG. 5.
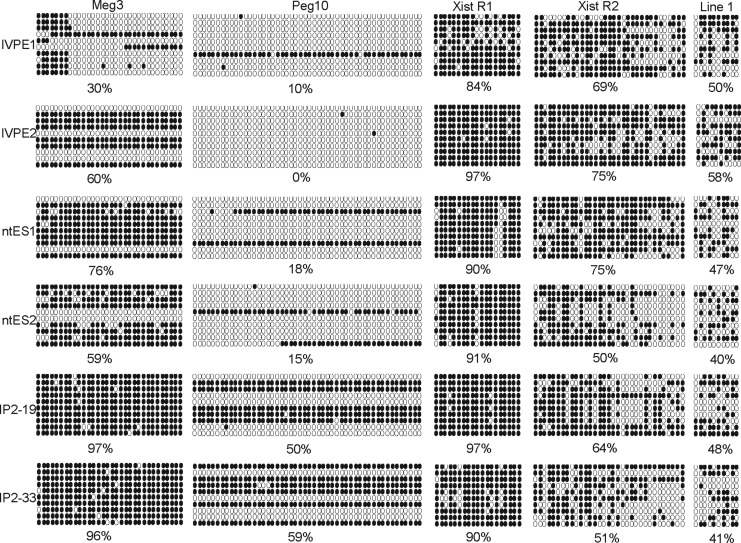
Bisulfate analysis of locus-specific DNA methylation. Meg3, Peg10, R1, and the R2 loci of Xist, repetive sequence Line 1 were detected. (Filled circles) Methylated CpGs; (open circles) unmethylated CpGs. Each row in each cell line represents each sequenced clone of each cell line. Percentage of methylation is an average of 10 clones for each cell line.
Locus-specific analysis of DNA methylation by bisulfite sequencing
To further investigate differences in DNA methylation among the six pluripotent cell lines, we analyzed nine specific loci of eight genes within the genome: Oct4, Peg3, Peg10, H19, Igf2r, and Meg3 from the Dlk1–Dio3 imprinting cluster, the R1 and R2 loci of the Xist gene, and the repeating sequence Line 1. The POU domain transcription factor Oct4 is a critical regulator of pluripotency in the mammalian embryo. It is highly expressed in the ICM of the blastocyst (Lavial al., 2007). The two iPSC lines showed higher degrees of methylation (24% and 30%) in the Oct4 region than the other four cell lines, suggesting that levels of Oct4 expression might have been low in the iPSC lines. Bisulfite sequencing showed that methylation levels of Peg3 and Peg10 differed significantly among the six cell lines, ranging from 0% to 59%. H19 and Meg3 of Dlk1–Dio3 showed vastly different levels of DNA methylation.
Xist transcription contributes to inactivation of the X chromosome (Young et al., 2012). This chromosome is controlled by methylation of the Xist gene's minimal promoter. X chromosome reactivation has been found to be an important event in successful reprogramming (Maherali et al., 2007). Long interspersed nuclear element-1 (Line-1 or L1) retrotransposons account for nearly 17% of human genomic DNA. They are a major evolutionary force and have reshaped the structure and function of the mammalian genome (Beck et al., 2010). Neither the R1 and R2 loci of the Xist gene nor repeating sequence Line-1 differed significantly with respect to the degree of DNA methylation among the six cell lines.
Discussion
ICM-derived ESCs are universally recognized as the best pluripotent stem cells currently available (Robinton and Daley, 2012). Application of and further research on these cells, however, are severely constrained for security reasons, specifically the fact that they can be obtained only from the ICM of developing blastocysts. Scientists have endeavored to create substitutes for ESCs in the form of reprogrammed somatic cells. This led to the invention of two vital techniques—ntESCs and iPSCs (Yamanaka and Blau, 2010). Both techniques are capable of reprogramming somatic cells into pluripotent stem cells that can develop into viable individuals. Mouse iPSCs do not require the use any oocytes, thereby circumventing ethical restrictions, which makes them an asset in the field of regenerative biology (Yamanaka, 2008). Unfortunately, the epigenetic characteristics of these cell types are poorly understood, so comparing epigenetic differences among iPSCs, ntESCs, and ESCs is of critical importance to the identification and development of suitable applications for iPSCs.
In this article, we have compared expression levels of enzymes involved in epigenetic modifications across six pluripotent stem cell lines. Real-time PCR showed that all examined enzymes had different levels of expression in these six cell lines. Out of the 11 genes evaluated, six (Dnmt3a, Dnmt3b, Tet1, Ezh2, Mll1, and Lsd1) showed abnormally low levels of expression in the two iPSC lines. To explain these results, we used reverse-phase HPLC to analyze methylation levels of genomic DNA. This analysis showed that the ntESC lines to have lower genomic 5mC content than the iPSC lines, indicating that ntESCs had been reprogrammed more successfully than iPSCs.
After evaluating the results of the quantitative PCR and reverse-phase HPLC analyses, we tried to discern a correlation between genomic 5mC content and the expression levels of DNA methylation and demethylation enzymes. No such correlations were apparent. Although iPS-19 and of iPS-33 showed high levels of genomic 5mC, they also showed relatively low degrees of expression of demethylation-related enzymes Dnmt1, Dnmt3a, and Dnmt3b. The level of Tet1 expression varied wildly across different strains, but Tet3 and Aid/Aicda were hardly expressed at all. These results are consistent with those of previous studies (data not shown). This shows that incomplete reprogramming of somatic cells caused abnormally high levels of genomic 5mC in the iPSC lines, although no direct evidence was collected here. Because the iPSCs were from chimeric mice, these results also suggest that the iPSCs remained incompletely reprogrammed despite their germ-line chimeric abilities.
To determine where the incomplete reprogramming of the iPSCs was concentrated, we conducted locus-specific analysis of DNA methylation on nine loci. Green fluorescence encoded by GFP in the promoter region of Oct4 (i.e., the expression of GFP driven by the Oct4 promoter) indicated that Oct4 was expressed in germ-line chimeric iPSCs (Fig. 1E), even though the expression levels were relatively low compared to those of the ESCs and ntESCs (Fig. 2). The incomplete reprogramming of Oct4 promoter region DNA methylation was reflected in the low methylation levels, specifically 24% and 30.0%. The Xist and Line-1 repeating sequences differed relatively little in methylation levels, but Peg3, Peg10, and H19 exhibited high degrees of variation in methylation levels. The Meg3 Dlk1–Dio3 imprinting cluster was incompletely methylated in ESCs and ntESCs, suggesting maternal expression of the Meg3 gene in these cell lines. In germ-line chimeric iPSCs, Meg3 was almost entirely methylated. In the ESC and ntESC lines, expression of Meg 3 was at least twice as high as in the iPSC lines (Fig. 2). This is consistent with previously published studies, which showed the Dlk1–Dio3 region to be activated in fully pluripotent mouse stem cells but almost completely repressed in partially pluripotent cells (Stadtfeld et al., 2010; Ito et al., 2011; Stadtfeld et al., 2012).
Previous studies show that continuous passaging of iPSCs abrogates transcriptional, epigenetic, and functional differences (Nishino et al., 2011). However, our study indicates that, after 30 passages in vitro, the two iPSC lines still differed significantly from the other pluripotent cell lines with respect to epigenetic modification.
The results of this study indicate that germ-line chimeric iPSCs remain incompletely reprogrammed and that Oct4 gene expression might be not the best indicator of complete reprogramming of somatic cells. Therefore, it is necessary to conduct further studies that evaluate the epigenetic differences of germ-line chimeric iPSCs and iPSCs with successful tetraploid complementation.
Supplementary Material
Acknowledgments
This work was supported by research grants from the National Natural Science Foundation of China (no. 31271597), National Basic Research Program of China (no. 2011CBA01000), and National Transgenic Breeding Program of China (no. 2011ZX08008-002).
Author Disclosure Statement
No competing financial interests exist.
References
- Adamo A. Sese B. Boue S. Castano J. Paramonov I. Barrero M.J. Belmonte J.C.I. Molecular mechanisms and potential functions of histone demethylases. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Beck C.R. Collier P. Macfarlane C. Malig M. Kidd J.M. Eichler E.E. Badge R.M. Moran J.V. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien K.R. Regenerative medicine and human models of human disease. Nature. 2008;453:302–305. doi: 10.1038/nature07037. [DOI] [PubMed] [Google Scholar]
- Chin M.H. Mason M.J. Xie W. Volinia S. Singer M. Peterson C. Ambartsumyan G. Aimiuwu O. Richter L. Zhang J. Khvorostov I. Ott V. Grunstein M. Lavon N. Benvenisty N. Croce C.M. Clark A.T. Baxter T. Pyle A.D. Teitell M.A. Pelegrini M. Plath K. Lowry W.E. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho D.D. You J.S. Jones P.A. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J. Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A. Alajem A. Meshorer E. Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E.L. Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K. Jaenisch R. Nuclear transplantation, embryonic stem cells, and the potential for cell therapy. N Engl J Med. 2003;349:275–286. doi: 10.1056/NEJMra035397. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K. Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- Hussein S.M. Batada N.N. Vuoristo S. Ching R.W. Autio R. Narva E. Ng S. Sourour M. Hamalainen R. Olsson C. Lundin K. Mikkola M. Trokovic R. Peitz M. Brustle O. Bazett-Jones D.P. Alitalo K. Lahesmaa R. Nagy A. Otonkoski T. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Ito S.C. D'Alessio A. Taranova O.V. Hong K. Sowers L.C. Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2011;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Doi A. Wen B. Ng K. Zhao R. Cahan P. Kim J. Aryee M.J. Ji H. Ehrlich L.I.R. Yabuuchi A. Takeuchi A. Cunniff K.C. Hongguang H. McKinney-Freeman S. Naveiras O. Yoon T.J. Irizarry R.A. Jung N. Seita J. Hanna J. Murakami P. Jaenisch R. Weissleder R. Orkin S.H. Weissman I.L. Feinberg A.P. Daley G.Q. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Zhao R. Doi A. Ng K. Unternaehrer J. Cahan P. Hongguang H. Loh Y.-H. Aryee M.J. Lensch M.W. Li H. Collins J.J. Feinberg A.P. Daley G.Q. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotech. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra S.M. Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Lavial F. Acloque H. Bertocchini F. Macleod D.J. Boast S. Bachelard E. Montillet G. Thenot S. Sang H.M. Stern C.D. Samarut J. Pain B. The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development. 2007;134:3549–3563. doi: 10.1242/dev.006569. [DOI] [PubMed] [Google Scholar]
- Lister R. Pelizzola M. Kida Y.S. Hawkins R.D. Nery J.R. Hon G. Antosiewicz-Bourget J. O'Malley R. Castanon R. Klugman S. Downes M. Yu R. Stewart R. Ren B. Thomson J.A. Evans R.M. Ecker J.R. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N. Sridharan R. Xie W. Utikal J. Eminli S. Arnold K. Stadtfeld M. Yachechko R. Tchieu J. Jaenisch R. Plath K. Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Nishino K. Toyoda M. Yamazaki-Inoue M. Fukawatase Y. Chikazawa E. Sakaguchi H. Akutsu H. Umezawa A. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7:e1002085. doi: 10.1371/journal.pgen.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M. Bell D.W. Haber D.A. Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Polo J.M. Liu S. Figueroa M.E. Kulalert W. Eminli S. Tan K.Y. Apostolou E. Stadtfeld M. Li Y. Shioda T. Natesan S. Wagers A.J. Melnick A. Evans T. Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotech. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton D.A. Daley G.Q. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. Shilatifard A. The chromatin signaling pathway: Diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M. Apostolou E. Akutsu H. Fukuda A. Follett P. Natesan S. Kono T. Shioda T. Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M. Apostolou E. Ferrari F. Choi J. Walsh R.M. Chen T. Ooi S.S. Kim S.Y. Bestor T.H. Shioda T. Park P.J. Hochedlinger K. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44:398–405. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M. Koh K.P. Shen Y. Pastor W.A. Bandukwala H. Brudno Y. Agarwal S. Iyer L.M. Liu D.R. Aravind L. Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson J.A. Itskovitz-Eldor J. Shapiro S.S. Waknitz M.A. Swiergiel J.J. Marshall V.S. Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Whyte W.A. Bilodeau S. Orlando D.A. Hoke H.A. Frampton G.M. Foster C.T. Cowley S.M. Young R.A. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Pluripotency and nuclear reprogramming. Phil Trans R Soc B: Biol Sci. 2008;363:2079–2087. doi: 10.1098/rstb.2008.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Blau H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.A. Larson D.E. Sun C.-W. George D.R. Ding L. Miller C.A. Lin L. Pawlik K.M. Chen K. Fan X. Schmidt H. Kalicki-Veizer J. Cook L.L. Swift G.W. Demeter R.T. Wendl M.C. Sands M.S. Mardis E.R. Wilson R.K. Townes T.M. Ley T.J. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.-y. Li W. Lv Z. Liu L. Tong M. Hai T. Hao J. Guo C.-l. Ma Q.-w. Wang L. Zeng F. Zhou Q. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.