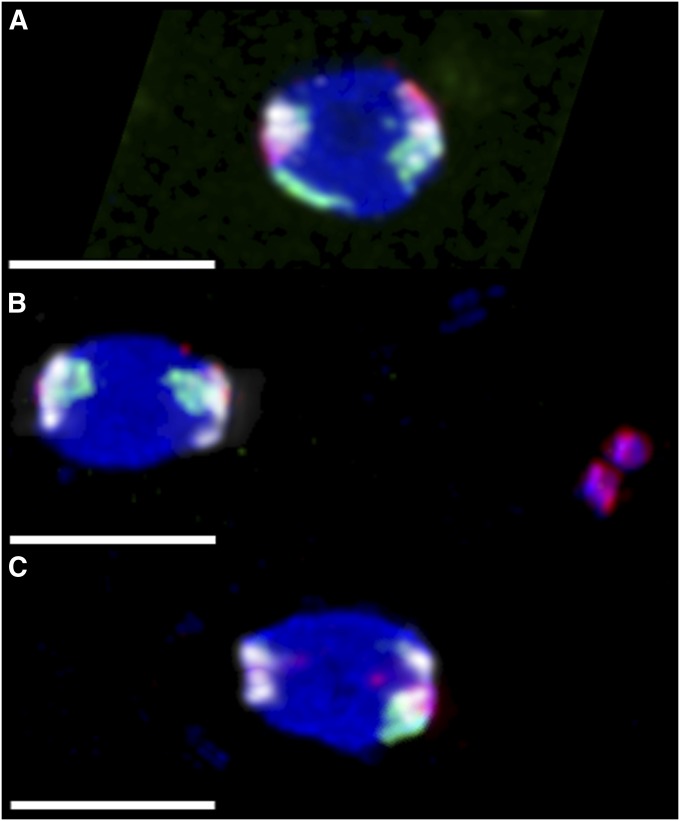
Figure 4 .
Chromosome loss in nod mutant oocytes. Experimental virgin females were aged 4 dpe with yeast and no males, and then FISH labeled with probes that predominantly hybridized to the X (green), 4 (red), and 2L–3L (white). As chiasmate chromosomes do not undergo meiotic errors in nod, the two pairs of white 2L–3L foci were used to infer the direction of the meiotic spindles, which are positioned horizontally. Three configurations are shown here, out of 20 possible configurations, not all of which were actually observed (Table S5). All bars, 4 µm. (A) An oocyte from a heterozygous control y w/y w noda; pol female, showing biorientation of both X and 4. This oocyte would always produce a euploid oocyte; 200/200 control oocytes scored showed this proper biorientation. (B) An oocyte from a homozygous y w noda; pol female, showing the two X homologs correctly bioriented, while the two 4 homologs are together off of the spindle to the right. This X⇔X oocyte would always produce normal-X, nullo-4 progeny. (C) An oocyte from a homozygous y w noda; pol female, showing the left pole with no X or 4, and single X and 4 chromosomes associated with the right pole; the other X and 4 homologs were visible elsewhere in the ooplasm (not shown). If the left spindle pole became the egg pronucleus, this Ø⇔X4 oocyte would produce a nullo-X, nullo-4 progeny, while the right pole would result in euploid progeny.