Abstract
Cell-cycle progression is monitored by checkpoint pathways that pause the cell cycle when stress arises to threaten the integrity of the genome. Although activation of checkpoint pathways has been extensively studied, our understanding of how cells resume the cell cycle when the stress is resolved is relatively limited. In this study, we identify the Saccharomyces cerevisiae F-box protein Dia2 as a novel player in the S-phase checkpoint recovery pathway. Dia2 is required for robust deactivation of the Rad53 checkpoint kinase and timely completion of DNA replication during recovery from DNA damage induced by methyl methanesulfonate (MMS). Aiming to identify the substrate of SCFDia2 (Skp1/Cul1/F-box Dia2) in checkpoint recovery, we performed a genetic screen to identify suppressors of dia2Δ cells. The screen identified a new checkpoint-defective allele of MRC1 truncated at the C terminus. We found that checkpoint-defective mrc1 alleles suppress the MMS sensitivity and the checkpoint recovery defect of dia2Δ cells. In addition, Dia2 contributes to Mrc1 degradation during S-phase checkpoint recovery. Furthermore, induced degradation of checkpoint-functional Mrc1 partially rescues the checkpoint recovery defect of dia2Δ cells. We propose a model in which Dia2 mediates Mrc1 degradation to help cells resume the cell cycle during recovery from MMS-induced DNA damage in S-phase.
Keywords: Dia2, Mrc1, DNA damage, checkpoint recovery
THE cell division cycle is tightly regulated to preserve genomic integrity and the viability of cells. Cells constantly monitor cell-cycle progression and employ checkpoints to pause the cell cycle when genome maintenance is threatened by genotoxins (Weinert and Hartwell 1988; Hartwell and Weinert 1989; Elledge 1996). For example, the S-phase checkpoint slows down DNA replication in the face of DNA damage while repair pathways are activated to resolve the damage (Rhind and Russell 2000). Any unrepaired damage in the newly synthesized DNA will trigger the G2/M DNA damage checkpoint to prevent cells from segregating the genetic material before the DNA damage is resolved (Weinert and Hartwell 1988; O’Connell et al. 2000; Rhind and Russell 2000).
During DNA replication, cells monitor the accumulation of single-strand DNA as a result of replication stress or DNA damage to activate the S-phase checkpoint (Costanzo et al. 2003; Zou and Elledge 2003; Fanning et al. 2006; Cimprich and Cortez 2008). In the budding yeast Saccharomyces cerevisiae, the initial signaling of the S-phase checkpoint leads to activation and recruitment of the Mec1/ATR kinase to the region of stress or damage (Kondo et al. 2001; Melo et al. 2001; Osborn and Elledge 2003). From there, Mec1 relays the checkpoint signal to downstream effectors through mediators that include Mrc1, Rad9, Tof1, and Csm3 (Navas et al. 1996; Vialard et al. 1998; Alcasabas et al. 2001; Foss 2001; Tong et al. 2004). In the case of Mrc1 and Rad9, these mediators are subjected to phosphorylation at Mec1 consensus S/TQ sites, which in turn facilitates the recruitment of a key downstream effector, the Rad53 kinase (Sun et al. 1998; Vialard et al. 1998;Alcasabas et al. 2001; Gilbert et al. 2001; Schwartz et al. 2002; Osborn and Elledge 2003). Once recruited, Rad53 is activated by Mec1 phosphorylation and autophosphorylation in trans (Vialard et al. 1998; Pellicioli et al. 1999; Sweeney et al. 2005; Chen and Zhou 2009). In the case of Mrc1, in addition to these S/TQ sites, other residues are also required to efficiently mediate checkpoint activation (Naylor et al. 2009). With the activation of Rad53 by the S-phase checkpoint, cells stabilize the replication fork and prevent origins from firing inappropriately (Santocanale and Diffley 1998; Shirahige et al. 1998; Tercero and Diffley 2001; Sogo et al. 2002; Branzei and Foiani 2005).
As important as it is for cells to activate the S-phase checkpoint in the face of DNA damage, cells must deactivate the checkpoint to resume the cell cycle after exposure to the DNA damage in a process termed checkpoint recovery (Van Vugt and Medema 2004; Bartek and Lukas 2007). Two previous studies provided evidence that in budding yeast, Rad53 dephosphorylation by phosphatases Pph3 and Ptc2 is required for recovery from MMS-induced DNA damage in S-phase (O’Neill et al. 2007; Szyjka et al. 2008). Indeed, Rad53 dephosphorylation is sufficient for fork restart during checkpoint recovery (Szyjka et al. 2008). Interestingly, fork recovery from replication stress agent hydroxyurea (HU) is not dependent on the Rad53 phosphatases (Travesa et al. 2008). Rather, fork recovery from HU is dependent on the chromatin remodeling complex Ino80 (Shimada et al. 2008).
We recently identified a previously uncharacterized linkage between the replication stress response and the SCF ubiquitin–proteasome pathway (Kile and Koepp 2010), a system that is better known for its role in protein turnover during cell-cycle progression (Ang and Harper 2005). An SCF ubiquitin ligase complex consists of Skp1, Cul1, Rbx1, and an F-box protein, which provides specificity of the complex (Feldman et al. 1997; Skowyra et al. 1997; Deshaies 1999; Kamura et al. 1999). Interestingly, we found that the proteolysis of the S. cerevisiae F-box protein Dia2 is regulated by the S-phase checkpoint. Indeed, Dia2 is highly stabilized when the checkpoint is activated in the presence of MMS (Kile and Koepp 2010). Furthermore, dia2 null (dia2Δ) cells are sensitive to MMS-induced DNA damage (Blake et al. 2006; Koepp et al. 2006). These findings suggest that Dia2 plays a role in the S-phase checkpoint. Because Rad53 is constitutively phosphorylated in the absence of Dia2 (Pan et al. 2006), it seems unlikely that Dia2 is required for checkpoint activation. Consistent with the data showing hyperactivation of Rad53 in dia2Δ cells, DNA replication is slow in dia2Δ cells in the presence of MMS (Blake et al. 2006).
The checkpoint mediator Mrc1 has recently been identified as a ubiquitin-mediated degradation substrate of SCFDia2 (Mimura et al. 2009). In addition to its role in checkpoint activation, Mrc1 also travels with the replication fork and is required for efficient DNA replication in an unperturbed S-phase (Osborn and Elledge 2003; Szyjka et al. 2005). The degradation of Mrc1 is most prominent in S-phase cells arrested in HU (Mimura et al. 2009). However, it remains an open question what the biological relevance of Mrc1 degradation is and whether Mrc1 is degraded for a role in an unperturbed S-phase or in response to the S-phase checkpoint activation. Intriguingly, the human homolog of Mrc1, Claspin, is targeted for proteasome degradation by the SCFβ-TrCP complex during recovery from replication stress or DNA damage before mitotic entry (Mailand et al. 2006; Peschiaroli et al. 2006). This degradation is regulated by Polo-like kinase-1 (Plk1) phosphorylation, which precedes the interaction between SCFβ-TrCP and Claspin (Mamely et al. 2006; Peschiaroli et al. 2006).
To better understand the function of Dia2 in the S-phase checkpoint, we performed a genetic screen to identify potential substrates of Dia2 and we investigated a possible role for Dia2 in checkpoint recovery from MMS-induced DNA damage.
Materials and Methods
Yeast cultures and cell cycle
Yeast cultures were grown according to standard protocols (Rose et al. 1990). For checkpoint activation experiments, cells were arrested using α-factor (GenScript) and then transferred into yeast peptone dextrose (YPD) containing 0.033% MMS. For checkpoint recovery experiments, cells were arrested by α-factor and then transferred into YPD containing 0.033% MMS for 40 min prior to transferring into YPD without MMS. Nocodazole (Sigma) was added to a final concentration of 15 μg/ml.
Genetic suppressor screen
dia2Δ spontaneous suppressors were selected on media containing 0.007% MMS. MMS-resistant candidates (105) were backcrossed to the dia2Δ strain to identify 9 recessive mutants, at least 3 of which resulted from a distinct, single-gene mutant. Mutants that exhibited higher MMS resistance than wild type in a DIA2 background were eliminated. The three single-hit, recessive candidates were crossed to the mrc1Δ and ctf4Δ strains for verification purposes.
Plasmid and strain construction
Tables 1, 2, and 3 contain a list of strains, plasmids, and oligonucleotides, respectively, used in this study. Deletion strains were generated by standard PCR replacement approaches (Rose et al. 1990). The following oligonucleotide pairs were used to make the deletion strains used in this study: CMF024–CMF025 (mrc1Δ::HIS3), CMF084–CMF085 (csm3Δ::KanMX), CMF086–CMF087 (rad9Δ::KanMX), CMF091–CMF092 (pph3Δ::KanMX), CMF103–CMF104 (tof1Δ::URA3). To construct pCMF001, the MRC1 locus was amplified with primers CMF013 and CMF014 and ligated to pRS415 (Sikorski and Hieter 1989) using XhoI. To generate pCMF002 (mrc1P263A) and pCMF003 (mrc1Q966stop), the suppressor mrc1 allele was amplified from the genomic DNA of the mrc1 suppressor strain. NdeI–NheI and NheI–PacI regions of pCMF001 were replaced with those of the suppressor allele to generate pCMF002 and pCMF003, respectively. pCMF011 (mrc1S965A) was constructed using primer sets CMF014–CMF018 (5′ fragment) and CMF017–CMF013 (3′ fragment). The fragments were combined by PCR before ligating to pRS415 using XhoI. pCMF013 (mrc11-971) was made in a similar fashion using primer sets CMF014–CMF019 and CMF021–CMF013, generating a stop codon mutation at residue 972.
Table 1 . Strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| Y80 | can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 MATa | Koepp et al. (2006) |
| DKY194 | As Y80 but dia2Δ::KanMX | Koepp et al. (2006) |
| AKY188 | As Y80 but dia2Δ::KanMX::9MYC-DIA2-ΔF (bp Δ670-792) URA3 | Kile and Koepp (2010) |
| DKY812 | As Y80 but pph3Δ::KanMX | This study |
| DKY826 | As Y80 but dia2Δ::KanMX pph3Δ::KanMX | This study |
| DKY643 | As Y80 but mrc1Δ::HIS3 | This study |
| DKY645 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3 | This study |
| DKY669 | As Y80 but mrc1Δ::HIS3::mrc11-971 LEU2 | This study |
| DKY781 | As Y80 but rad9Δ::KanMX | This study |
| DKY820 | As Y80 but tof1Δ::URA3 | This study |
| DKY786 | As Y80 but csm3Δ::KanMX | This study |
| DKY782 | As Y80 but rad9Δ::KanMX mrc1Δ::HIS3::mrc11-971 LEU2 | This study |
| DKY852 | As Y80 but tof1Δ::KanMX mrc1Δ::HIS3::mrc11-971 LEU2 | This study |
| DKY800 | As Y80 but csm3Δ::KanMX mrc1Δ::HIS3::mrc11-971 LEU2 | This study |
| DKY728 | As Y80 but mrc1Δ::HIS3::3HA-MRC1 LEU2 RAD53::RAD53-3FLAG TRP1 | This study |
| DKY729 | As Y80 but mrc1Δ::HIS3::3HA-mrc11-971 LEU2 RAD53::RAD53-3FLAG TRP1 | This study |
| DKY783 | As Y80 but rad9Δ::KanMX mrc1Δ::HIS3::3HA-MRC1 LEU2 RAD53::RAD53-3FLAG TRP1 | This study |
| DKY784 | As Y80 but rad9Δ::KanMX mrc1Δ::HIS3::3HA-mrc11-971 LEU2 RAD53::RAD53-3FLAG TRP1 | This study |
| Y2298 | can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 HIS::mrclAQ-MYC13 MATa | Osborn and Elledge (2003) |
| DKY769 | As Y80 but dia2Δ::KanMX HIS::mrclAQ-MYC13 | This study |
| DKY672 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3::mrc11-971 LEU2 | This study |
| DKY824 | As Y80 but pph3::KanMX mrc1Δ::HIS3::mrc11-971 LEU2 | This study |
| DKY765 | As Y80 but HIS::mrclAQ-MYC13 RAD53::RAD53-3FLAG TRP1 | This study |
| DKY730 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3::3HA-MRC1 LEU2 RAD53::RAD53-3FLAG TRP1 | This study |
| DKY731 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3::3HA-mrc11-971 LEU2 RAD53::RAD53-3FLAG TRP1 | This study |
| DKY802 | As Y80 but dia2Δ::KanMX HIS::mrclAQ-MYC13 RAD53::RAD53-3FLAG TRP1 | This study |
| DKY818 | As Y80 but dia2Δ::KanMX rad9::KanMX | This study |
| DKY854 | As Y80 but dia2Δ::KanMX tof1::URA3 | This study |
| DKY816 | As Y80 but dia2Δ::KanMX csm3Δ::KanMX | This study |
| DKY688 | As Y80 but mrc1Δ::HIS3::3HA-MRC1 LEU2 | This study |
| DKY698 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3::3HA-MRC1 LEU2 | This study |
| DKY919 | As Y80 but RAD9::RAD9-3FLAG LEU2 bar1Δ::URA3 | This study |
| DKY920 | As Y80 but dia2Δ::KanMX RAD9::RAD9-3FLAG LEU2 bar1Δ::URA3 | This study |
| DKY710 | As Y80 but TOF1::TOF1-3FLAG TRP1 CSM3::CSM3-3MYC TRP1 | This study |
| DKY756 | As Y80 but dia2Δ::KanMX TOF1::TOF1-3FLAG TRP1 CSM3::CSM3-3MYC TRP1 | This study |
| DKY972 | As Y80 but mrc1Δ::HIS3::3HA-mrc1AQ LEU2 | This study |
| DKY973 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3::3HA-mrc1AQ LEU2 | This study |
| DKY862 | As Y80 but rpn4Δ::HIS3 pdr5Δ::LEU2 mrc1::HIS3::3HA-MRC1 LEU2 | This study |
| DKY946 | As Y80 but mrc1Δ::HIS3::3HA-mrc1Δ380-430,701-800 LEU2 | This study |
| DKY928 | As Y80 but mrc1Δ::HIS3::3HA-mrc1Δ461-557,701-800 LEU2 | This study |
| DKY949 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3::3HA-mrc1Δ380-430,701-800 LEU2 | This study |
| DKY931 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3::3HA-mrc1Δ461-557,701-800 LEU2 | This study |
| DKY914 | As Y80 but mrc1Δ::HIS3::3HA-mrc13SA LEU2 | This study |
| DKY970 | As Y80 but mrc1Δ::HIS3::3HA-AID3-111-MRC1 LEU2 ura3-1::ADH1-OsTIR1-9MYC URA3 | This study |
| DKY967 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3::3HA-AID3-111-MRC1 LEU2 ura3-1::ADH1-OsTIR1-9MYC URA3 | This study |
| DKY974 | As Y80 but dia2Δ::KanMX mrc1Δ::HIS3::3HA-AID3-111-mrc11-971 LEU2 ura3-1::ADH1-OsTIR1-9MYC URA3 | This study |
Table 2 . Plasmids used in this study.
| Plasmid | Relevant features | Reference |
|---|---|---|
| pRS415 | CEN LEU2 Ampr | Sikorski and Hieter (1989) |
| pCMF001 | MRC1 genomic locus (22591–17555 on chromosome III) in pRS415 | This study |
| pCMF002 | mrc1P263A in pRS415 | This study |
| pCMF003 | mrc1Q966stop in pRS415 | This study |
| pCMF011 | mrc1S965A in pRS415 | This study |
| pCMF013 | mrc11-971 in pRS415 | This study |
| pCMF021 | mrc11-971 in pRS405 | This study |
| pCMF022 | 3HA-MRC1 in pRS405 | This study |
| pCMF023 | 3HA-mrc11-971 in pRS405 | This study |
| pCMF026 | TOF1-3FLAG C-terminal fragment in pRS404 | This study |
| pCMF027 | CSM3-3MYC C-terminal fragment in pRS404 | This study |
| pCMF028 | RAD53-3FLAG C-terminal fragment in pRS404 | This study |
| pCMF029 | RAD9-3FLAG C-terminal fragment in pRS405 | This study |
| pCMF041 | 3HA-mrc13SA in pRS405 | This study |
| pCMF045 | 3HA-mrc1Δ461-557,701-800 in pRS405 | This study |
| pCMF050 | 3HA-mrc1Δ380-430,701-800 in pRS405 | This study |
| pCMF053 | 3HA-AID3-111-MRC1 in pRS405 | This study; derived from pMK38 Nishimura et al. (2009) |
| pCMF054 | 3HA-AID3-111-mrc11-971 in pRS405 | This study |
| pCMF055 | 3HA-mrc1AQ in pRS405 | This study; derived from pAO138 Osborn and Elledge (2003) |
Table 3 . Oligonucleotides used in this study.
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| CMF010 | GGACACCAACTCTACTGGCTC |
| CMF013 | CCCCTCGAGAACGCATAGAAGACTTGGTTCG |
| CMF014 | CCCCTCGAGAATTGAAAGTGGTGAGTATTTC |
| CMF017 | GTCATTCACAAATGCTCAAACTGATTC |
| CMF018 | GAATCAGTTTGAGCATTTGTGAATGAC |
| CMF019 | GATAAAAAACCAGTTTACTGTTTTTCAAGTGGTCGAATC |
| CMF021 | GAAAAACAGTAAACTGGTTTTTTATCTTTTCCGAAG |
| CMF024 | GAAGTTCGTTATTCGCTTTTGAACTTATCACCAAATATTTTAGTGGGCCTCCTCTAGTACACTC |
| CMF025 | AGCTTCTGGAGTTCAATCAACTTCTTCGGAAAAGATAAAAAACCAGCGCGCCTCGTTCAGAATG |
| CMF043 | GAATTTGTTTCCTGCTAGCTTTC |
| CMF044 | GTCGTATGGGTAACCTGCCATCACTAAAATATTTGGT |
| CMF045 | ACCAAATATTTTAGTGATGGCAGGTTACCCATACGAC |
| CMF046 | CAAAGCATGCAAGGCATCATCGGAACGTAGAGAAGCGTAATC |
| CMF047 | GATTACGCTTCTCTACGTTCCGATGATGCCTTGCATGCTTTG |
| CMF061 | CCCCTCGAGGAAGAAGTTACTCCAAGATTTG |
| CMF063 | GGGCTCGAGGGGAAGGAGACGATGATTATG |
| CMF064 | TTTCTCGAGCGAGTTTTGGACGAACGTGGG |
| CMF065 | CCCCTCGAGCCCGTTGGTTATCGAAAATCG |
| CMF068 | GCCGCATAGCTCGAATCCCATAAAGCCCATTTCCTTCATAGC |
| CMF069 | GCTATGAAGGAAATGGGCTTTATGGGATTCGAGCTATGCGGC |
| CMF070 | TTATTACCTTCAATGACATTGCTAGCTACTATTAAGATCCTCCTC |
| CMF071 | GAGGAGGATCTTAATAGTAGCTAGCAATGTCATTGAAGGTAATAA |
| CMF072 | ATCTTTATAATCGAGCTCCAGATCATCACTATCACCTTGGCT |
| CMF073 | AGCCAAGGTGATAGTGATGATCTGGAGCTCGATTATAAAGAT |
| CMF074 | GGATTAATTACTACATATTCATTCCAGTTACTTGTCATCGTCATC |
| CMF075 | GATGACGATGACAAGTAACTGGAATGAATATGTAGTAATTAATCC |
| CMF077 | AGGTTGTCTCGAGTTCATTGCTTC |
| CMF078 | TTTATAATCGAGCTCCAGCTTCGAAAATTGCAAATTCTCGGGGCC |
| CMF079 | GGCCCCGAGAATTTGCAATTTTCGAAGCTGGAGCTCGATTATAAA |
| CMF080 | CCCCTCGAGGTCCATAAATTCCTGCAGTTACTT |
| CMF081 | CCCCTCGAGCATCCGCTAGCTAAATCTTTAG |
| CMF082 | ATCTTTATAATCGAGCTCCAGTCTAACCTCAGAAATAGTGTTG |
| CMF083 | CAACACTATTTCTGAGGTTAGACTGGAGCTCGATTATAAAGAT |
| CMF084 | GATTAAAATGCCATGAAAACGTGAACAGAAACTTTTATTGAGGTCGTTTAGCTTGCCTCGTCCCCG |
| CMF085 | TAGATGCCCACACGCACGTTTGGATTATTACCTTCAATGACATTGTTAAGGGTTCTCGAGAGCTCG |
| CMF086 | CGCCATAGAAAAGAGCATAGTGAGAAAATCTTCAACATCAGGGCTGTTTAGCTTGCCTCGTCCCCG |
| CMF087 | AATCGTCCCTTTCTATCAATTATGAGTTTATATATTTTTATAATTTTAAGGGTTCTCGAGAGCTCG |
| CMF091 | AAGTAAAACAGCACGAAAAAAGTGATTACAAATTTCAAGGGAGATGTTTAGCTTGCCTCGTCCCCG |
| CMF092 | AAAAAAAGAAAAATGCACTTGACAATTAGAGTGCCTGTTAAAAATTTAAGGGTTCTCGAGAGCTCG |
| CMF103 | AGCTTGTGGGGTTTAGTGTATCTTTAATATAGGAGGGCGCACACTAGCTTTTCAATTCAATTCATC |
| CMF104 | CTAAAATTACACGTATTAAAGGGATTAATTACTACATATTCATTCCACACCGCATAGGGTAATAAC |
| CMF129 | CAGATTCAGCTCGACACTGCCGGAACGTAGAGAAGCGTAATC |
| CMF130 | GATTACGCTTCTCTACGTTCCGGCAGTGTCGAGCTGAATCTG |
| CMF135 | GGCGACTATATTAAACCTGAAGGCAAG |
| CMF136 | CGCCCATGATGCCGGTTCTGACGCAGGGTCAGAGGCTTCTGG |
| CMF137 | CCAGAAGCCTCTGACCCTGCGTCAGAACCGGCATCATGGGCG |
| CMF138 | GGAGAAAGAATAAGGGCATGAATGAAGAAC |
| CMF140 | GAGACAAGAATAAATGAGAAAAGGGTTCCAC |
| CMF141 | TTCATTACCACTCAAATTCGGCCTTTGAGACAACTTTTG |
| CMF142 | CAAAGGCCGAATTTGAGTGGTAATGAAATTGCCGATTATG |
| CMF143 | GCAAGATGCTTTGAATACAGAACTGCTG |
| CMF144 | GCTCTAAGCTTTCTTTCTGTTTTAGTTGCAATTTCTC |
| CMF145 | CTAAAACAGAAAGAAAGCTTAGAGCTAGAACTAAGTGATG |
| CMF158 | CTGAGAGATTGGCAAATTTATTCTTGTCTCATCAGTTAG |
| CMF159 | GAGACAAGAATAAATTTGCCAATCTCTCAGTTATCAAAG |
| CMF160 | CACTACCAGATGATTCATAATCGGC |
| CMF165 | CAAAGCATGCAAGGCATCATCCGCCGCCGCCTCCGGGCCACC |
| CMF166 | GGTGGCCCGGAGGCGGCGGCGGATGATGCCTTGCATGCTTTGTCC |
| CMF167 | CTTTCTGATGATCCAGAATTTGTTTCCTGC |
Alleles were integrated using standard homologous recombination approaches (Rose et al. 1990). To generate the mrc11-971 strain, the entire XhoI fragment of mrc11–971 was moved from pCMF013 to pRS405 to generate pCMF021. The plasmid was linearized with NdeI and transformed into the mrc1Δ strain, and the same is true for the integration of all other mrc1 alleles used in this study. For N-terminal-tagged MRC1 strains, fragments of MRC1 5′ UTR, the 3× HA epitope, and the MRC1 N terminus ORF (open reading frame) were amplified with primer sets CMF010–CMF044, CMF045–CMF046, and CMF047–CMF043, respectively. The products are combined by PCR and subcloned to pCMF001 and pCMF013 to generate 3HA–MRC1 and 3HA–mrc11-971. The XhoI fragments of 3HA–MRC1 and 3HA–mrc11-971 were subcloned to pRS405 to generate pCMF022 and pCMF023. 3HA–mrc13SA was generated by first amplifying the NheI–PacI region of MRC1 with primer sets CMF135–CMF137 and CMF136–CMF138. The fragments were combined by PCR and subcloned into pCMF022 to generate pCMF041. Generation of the mrc1 mutants deleted of the putative Dia2-binding regions were performed as follows: 5′ and 3′ fragments of MRC1 were amplified using two pairs of primers before the products were combined by PCR. Primer sets CMF010–CMF158 and CMF159–CMF160 were used to generate a mrc1Δ380-430 fragment, CMF140–CMF141 and CMF142–CMF143 were used to generate a mrc1Δ461-557 fragment, and CMF140–CMF144 and CMF145–CMF143 were used to generate a mrc1Δ701–800 fragment. The mrc1Δ701–800 fragment was first subcloned into pCMF022 using XbaI to generate 3HA–mrc1Δ701–800 in pRS405. The mrc1Δ380–430 fragment was then subcloned into 3HA–mrc1Δ701–800 using NdeI and NheI to generate pCMF050, whereas the mrc1Δ461–557 fragment was subcloned into 3HA–mrc1Δ701–800 using NdeI and XbaI to generate pCMF045. 3HA–mrc1AQ was constructed using an untagged mrc1AQ plasmid (pAO138 from Osborn and Elledge 2003). ApaI–PacI fragment of mrc1AQ was subcloned into pCMF022 to generate pCMF055.
MRC1 was conjugated to an auxin-inducible degron (AID) peptide containing amino acid residues 3–111 (Dreher et al. 2006; Nishimura et al. 2009). To generate the 3HA–AID3–111-MRC construct, 5′ UTR (containing 3xHA) and 5′ ORF of MRC1 were amplified using primer sets CMF10–CMF129 and CMF166–CMF167. The AID peptide was also amplified using primers CMF130 and CMF165. The three fragments were combined by PCR, and the product was digested with NdeI and NheI enzymes before subcloning into pCMF022 and pCMF023 to generate pCMF053 (3HA–AID3–111–MRC) and pCMF054 (3HA–AID3–111–mrc11-971), respectively.
Epitope-tagged TOF1 was generated by combining several fragments. The C-terminal TOF1 ORF, 3× FLAG, and 3′-UTR were amplified using primer sets CMF063–CMF072, CMF073–CMF074, and CMF075–CMF061. Those fragments were combined by PCR and ligated to pRS404 using XhoI to generate pCMF026. The plasmid was linearized with NsiI and transformed into the Y80 strain. A similar approach was used to generate CSM3–3MYC, using primer sets CMF064–CMF068 (ORF), CMF069–CMF070 (3× MYC), and CMF071–CMF065 (3′-UTR). Those fragments were combined by PCR and ligated to pRS404 using XhoI to generate pCMF027. The plasmid was linearized with NsiI and transformed into the Y80 strain. To construct RAD53–3FLAG, a C-terminal RAD53 fragment and 3× FLAG were amplified with primer sets CMF077–CMF078 and CMF079–CMF080. The fragments were combined by PCR and ligated to pRS404 using XhoI to generate pCMF028. The plasmid was linearized with SwaI and transformed into the Y80 strain. RAD9–3FLAG was generated in a similar fashion using primer sets CMF081–CMF082 (ORF) and CMF083–CMF080 (3× FLAG). The fragments were combined by PCR and ligated into pRS405 using XhoI to generate pCMF029. The plasmid was linearized with NsiI and transformed into a bar1Δ strain in Y80 background.
Growth and viability assays
Tenfold serial dilutions of 2 × 107 to 2 × 103 cells were spotted onto media using a replica plater. Plates were incubated at 30° for 2–3 days. For viability assays, cells were first grown in YPD to log phase. Equal numbers of cells were plated on YPD containing MMS. Plates were incubated at 30° for 4 days and CFU were counted. The percentage viability of cells was determined by dividing CFU on YPD with MMS by CFU on YPD without MMS.
Flow cytometry
Harvested cells were fixed with 70% ethanol and resuspended in 1× phosphate buffered saline (PBS). Cells were sonicated to break open clumps and subjected to RNase treatment (100 μg/ml) in Tris–EDTA overnight. Samples were stained with propidium iodide (Calbiochem) at a final concentration of 50 μg/ml in 1× PBS for 1 hr and analyzed by flow cytometry using FACSCalibur (BD Biosciences) and FlowJo software (Tree Star). The cell-cycle-distribution graphs were generated with Deltagraph (Red Rock). Quantification of 2C DNA content in each strain was achieved using flow cytometry data gated for a standard 2C distribution developed from an asynchronous cell population using FlowJo software (Tree Star). Percentages were calculated by dividing the number of cells with 2C DNA content by the total number counted. Average values were plotted in graphs with standard deviations used for error bars. P-values were calculated using paired Student’s t-test analysis.
Protein gel electrophoresis and Western blots
Protein extracts were prepared using 20% trichloroacetic acid (TCA) precipitation as previously described (Kile and Koepp 2010). For Rad53 deactivation experiments, protein samples were resolved in 6% Tris–glycine denaturing protein gels (SDS–PAGE). The very top modified band of Rad53 was quantified using ImageJ as a measure of Rad53 deactivation. For all other experiments, protein samples were resolved in 3–8% Tris-acetate gels (Invitrogen). 3HA–Mrc1, Csm3–3MYC, and Pgk1 were detected by anti-HA (Covance), anti-MYC 9E10 (Covance), and anti-Pgk1 (Molecular Probes) antibodies, respectively. Tof1–3FLAG, Rad9–3FLAG, and Rad53–3FLAG were detected by anti-FLAG (Sigma) antibody. Secondary antibody incubation, blot development, protein quantification, and loading normalization were performed as previously described (Kile and Koepp 2010).
Stability assays
Cells were grown in YPD to log phase before performing arrest and release checkpoint recovery experiments. Cycloheximide (CHX) (Sigma) was added to cell cultures to a final concentration of 200 μg/ml. For proteasome inhibitor experiments, the 3HA–MRC1rpn4Δ pdr5Δ strain was used and MG-132 (American Peptide) was added to the media at a final concentration of 50 μM.
Auxin-induced degradation of Mrc1
This system was adapted from Dreher et al. (2006) and Nishimura et al. (2009), provided by the Yeast Genetic Resource Center, Osaka University. During checkpoint recovery experiments, either vehicle (100% ethanol) or 1.5 mM indole-3-acetic acid (IAA) (Alfa Aesar) was added to cell cultures in YPD at 30°.
Results
Dia2 is required for effective checkpoint recovery from MMS-induced DNA damage
The mechanistic role of Dia2 in the S-phase checkpoint remains largely unknown. Because the Dia2 protein is stabilized when the S-phase checkpoint is activated in the presence of MMS (Kile and Koepp 2010) and the checkpoint remains active in dia2Δ cells (Pan et al. 2006), we hypothesized that the role of Dia2 lies downstream of checkpoint activation rather than at the initiation of signal. Furthermore, DNA replication in the presence of MMS was reported to be defective in dia2Δ cells (Blake et al. 2006). One possibility is that Dia2 is required for DNA replication to resume during checkpoint recovery. Thus, we asked if Dia2 plays a role in checkpoint recovery from MMS-induced DNA damage. Cells were arrested in late G1 by α-factor, released into media containing MMS to activate the S-phase checkpoint, and then released into media without MMS to observe DNA replication during checkpoint recovery. After cells had been released from media containing MMS, almost 70% of wild-type cells completed DNA replication in 60 min, whereas only 58% of dia2Δ cells completed DNA replication at the same time point (Figure 1A). Indeed, only at the 100-min time point did 70% of dia2Δ cells complete DNA replication. We did not observe any difference between wild-type and dia2Δ cells completing S-phase in the absence of MMS (Figure 1B), indicating that DNA replication is impeded during recovery from MMS-induced DNA damage but not in an unperturbed S-phase in dia2Δ cells. These results suggest that Dia2 has a role in checkpoint recovery.
Figure 1 .
Dia2 is required for checkpoint recovery from MMS-induced DNA damage in S-phase. (A) Cells were arrested in late G1 by α-factor (αF), released into rich media (YPD) + 0.033% MMS for 40 min, and then released into YPD. Samples were analyzed at the indicated time points by flow cytometry. 1C and 2C indicate DNA content. Percentage of cells with 2C DNA content is indicated on the right of selected profiles. (B) Cells were arrested in late G1 by αF and then released into YPD at 30°. Samples were analyzed by flow cytometry. (C and D) DIA2 genetically interacts with Rad53-phosphatase PPH3 in response to MMS. (C) Tenfold serial dilutions of the indicated strains were spotted on YPD or YPD + 0.007% MMS and incubated at 30°. (D) Equal numbers of cells were plated on media containing the indicated amounts of MMS, and colony-forming units were counted after 4 days at 30°. Error bars represent standard deviations from three independent experiments. (E and F) Dia2 functions in parallel to Pph3 for S-phase checkpoint recovery. Samples were prepared and analyzed as described in A and B.
We asked if Dia2 is required to form an SCF complex to regulate checkpoint recovery. We used the dia2ΔF-box mutant as this strain lacks only the F-box domain of Dia2, which is required for binding to Skp1 and therefore the rest of the SCF complex (Bai et al. 1996). As shown in Figure 1A, dia2ΔF-box cells completed DNA replication at a later time point than wild type. Indeed, the checkpoint recovery rate was similar between dia2Δ and dia2ΔF-box strains, with about 70% of cells completing DNA replication by 100 min. These data suggest that the ubiquitination function of Dia2 may be important for checkpoint recovery from MMS-induced DNA damage.
We asked where Dia2 fits in the checkpoint recovery pathway among players identified to have a role in the pathway. Previous studies showed that Rad53 is deactivated by phosphatases Pph3 and Ptc2 during S-phase checkpoint recovery (O’Neill et al. 2007; Szyjka et al. 2008). We generated a dia2Δ pph3Δ double-mutant strain and examined it for growth and viability on MMS-containing media. Serially diluted cells were spotted onto media with or without MMS to compare growth between wild-type, pph3Δ, dia2Δ, and dia2Δ pph3Δ strains (Figure 1C). To examine viability, equal numbers of cells were plated onto media containing various concentrations of MMS. The dia2Δ pph3Δ mutant exhibited both weaker growth and viability than the dia2Δ or the pph3Δ mutant on media containing MMS (Figure 1, C and D). We then examined checkpoint recovery with these strains. As expected, pph3Δ (O’Neill et al. 2007) and dia2Δ cells completed DNA replication at later time points than wild-type cells during checkpoint recovery (Figure 1E). Strikingly, the dia2Δ pph3Δ strain exhibited even slower checkpoint recovery compared to the single mutants. Indeed, the double mutant did not complete DNA replication by the last time point tested in this assay (Figure 1E). These data suggest that Dia2 plays an important role in S-phase checkpoint recovery and likely acts in parallel to the Rad53 phosphatase Pph3, although we cannot rule out that Dia2 and Pph3 may have a synthetic defect in fork progression from these data.
A genetic screen identified a checkpoint-defective mrc1 allele as a suppressor of dia2Δ
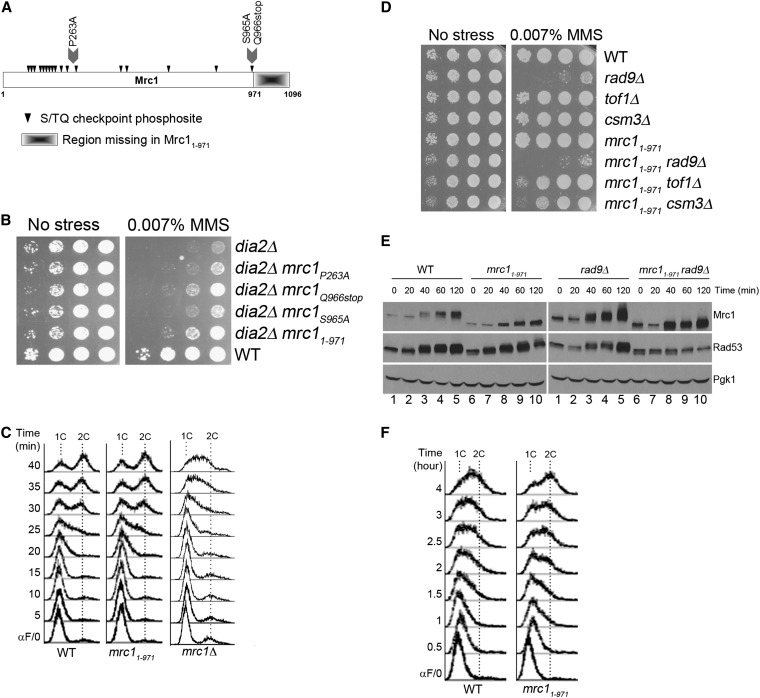
Our results suggested that Dia2 might target a specific protein for ubiquitin-mediated degradation during checkpoint recovery. As an unbiased approach to identifying potential targets, we began a genetic suppressor screen to identify genes involved in the same pathway as DIA2. As the dia2Δ mutant is hypersensitive to MMS, we screened for suppressors of this phenotype. We identified three single-hit, recessive mutants that partially suppressed the dia2Δ MMS sensitivity; these fell into three distinct complementation groups. With the recent identification of Mrc1 and Ctf4 as targets of SCFDia2 (Mimura et al. 2009), both of which have roles in the S-phase checkpoint, we crossed the suppressors to the mrc1Δ and the ctf4Δ strains to directly test for complementation. One of the mutants failed to complement mrc1Δ whereas all mutants complemented ctf4Δ. The identity of the remaining mutants remains to be determined. We isolated the allele of MRC1 (Figure 2A) and found that it contained a point mutation P263A and an early stop codon at Q966. We found that the early stop codon at Q966, but not the P263A point mutation, was responsible for the suppression (Figure 2B). Because the early stop codon is at the last SQ phosphosite of Mrc1, we investigated whether the last SQ site (S965A) or the C-terminal truncation (1–971) would suppress dia2Δ cells. We found that the mrc11-971 allele suppressed the dia2Δ strain’s MMS-sensitivity phenotype, whereas the mrc1S965A allele did not (Figure 2B).
Figure 2 .
A genetic screen identified a checkpoint-defective allele of mrc1 that suppresses the MMS sensitivity of dia2Δ. (A) Structural schematic of the Mrc1 protein. Arrowheads indicate S/TQ Mec1-directed phosphosites. Mutations used in these studies are marked. (B) mrc11–971 suppresses the MMS sensitivity of dia2Δ. The indicated strains were spotted using 10-fold serial dilutions on rich media with or without 0.007% MMS and incubated at 30°. (C) mrc11–971 is functional in DNA replication. Cells were arrested in G1 by α-factor and released into YPD at 30°. The indicated time points were analyzed by flow cytometry. 1C and 2C indicate DNA content. (D) The mrc11–971 allele exhibits negative genetic interactions with other S-phase checkpoint mediator mutants. Tenfold serial dilutions of the indicated strains were spotted on YPD or YPD + 0.007% MMS and incubated at 30°. (E) Checkpoint activation of Rad53 is compromised in mrc11–971. Cells were arrested in G1 by α-factor and released into YPD + 0.033% MMS at 30°. Protein samples were taken at the indicated time points. Pgk1 was used as a loading control. The checkpoint activation of Rad53 was measured using the intensity of Rad53 phosphorylation shift. (F) The mrc11–971 allele bypasses checkpoint-activated slowing of DNA replication. Samples were prepared as described in E and analyzed at the indicated time points by flow cytometry.
As Mrc1 has roles in both DNA replication (Szyjka et al. 2005) and checkpoint activation (Alcasabas et al. 2001; Osborn and Elledge 2003), we tested which function was required for suppression. A previous study has shown that deletion of residues beyond 988 do not compromise Mrc1 replication function (Naylor et al. 2009). To confirm functionality of DNA replication, wild-type and mrc11–971 cells were arrested in G1 and released into S-phase and DNA replication was monitored by flow cytometry. We found that the mrc11–971 strain progressed at the same rate as wild type through DNA replication, whereas the mrc1Δ control strain exhibited slow progression in S-phase (Figure 2C). We conclude that mrc11-971 is functional in DNA replication.
To examine the checkpoint function of the mrc11–971 strain, we tested (1) whether other checkpoint mediators are required for mrc11–971 to survive DNA damage, (2) the extent of Rad53 activation in response to MMS, and (3) if the checkpoint is active in mrc11–971 cells. First, the mrc11–971 mutant was crossed to checkpoint mediator mutants rad9Δ, tof1Δ, and csm3Δ, and single and double mutants were examined for sensitivity to MMS. The mrc11–971rad9Δ double mutant exhibited slightly weaker growth than the mrc11–971 or the rad9Δ single mutant on media containing MMS. A stronger negative genetic interaction was observed between mrc11–971 and tof1Δ, as well as between mrc11–971 and csm3Δ (Figure 2D). Second, we compared Rad53 activation in wild-type and mrc11–971 cells in response to MMS. Cells were arrested in G1 and released into media with MMS. Because Rad53 becomes phosphorylated during checkpoint activation (Pellicioli et al. 1999), the shift from nonphosphorylated to phosphorylated Rad53 was monitored over time in wild-type and mrc11–971 cells. Since Mrc1 and Rad9 both activate Rad53 in response to MMS (Vialard et al. 1998; Alcasabas et al. 2001; Gilbert et al. 2001; Osborn and Elledge 2003), we also tested Rad53 activation in the rad9Δ background. While the difference in Rad53 activation was subtle between wild-type and mrc11–971 cells, the Rad53 phosphorylation shift was mostly abolished in rad9Δ mrc11–971 cells (Figure 2E). This result indicates that Mrc11–971 is defective in activating Rad53 in response to MMS. Interestingly, the phosphorylation shift of Mrc11–971 is reduced compared to full-length Mrc1 (Figure 2E), even though the deletion mutant retains all of the S/TQ checkpoint phosphosites (Figure 2A). It is possible that the reduced phosphorylation of the Mrc11–971 protein may contribute to the reduced Rad53 activation in this strain, although the mechanism for how this is achieved is unclear. Last, we determined if the S-phase checkpoint is intact in mrc11–971 cells. If mrc11–971 is checkpoint defective, we expected that DNA replication in MMS would be faster in the mutant than wild type. Cells were treated as described for Figure 2E and DNA replication was monitored by flow cytometry. DNA replication within the first 2 hr was indistinguishable between wild-type and mrc11–971 cells. However, we noted a difference in the DNA replication profile between the two strains by the 3-hr time point. By 4 hr, the mrc11–971 strain completed DNA replication, whereas the majority of wild-type cells did not complete DNA replication (Figure 2F). These data indicate that this novel allele mrc11–971 is functional in DNA replication but partially defective in activating the S-phase checkpoint.
Checkpoint-defective mrc1 alleles suppress dia2Δ sensitivity to MMS and defects in checkpoint recovery
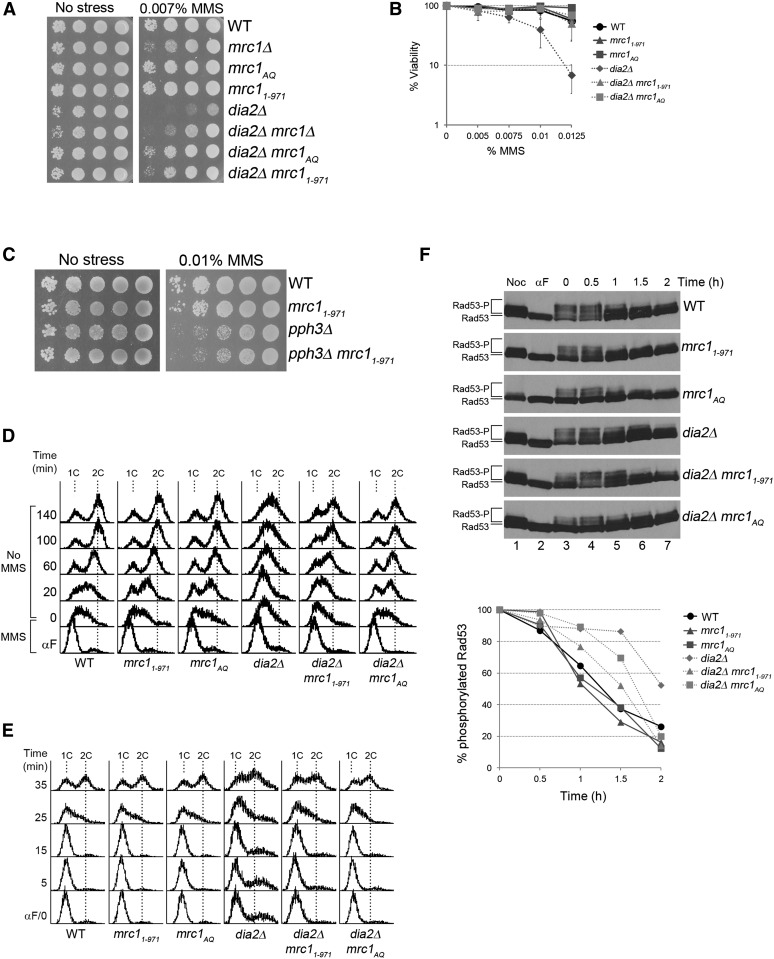
Given that Mrc1 was reported to be an ubiquitin-mediated degradation substrate of Dia2 (Mimura et al. 2009), our results raised the possibility that Dia2 mediates Mrc1 degradation for checkpoint recovery. In this case, we would predict that the absence of the substrate would suppress the MMS sensitivity of dia2Δ cells. To test this, we generated a dia2Δ mrc1Δ double mutant and examined it for growth on media with or without MMS. As shown in Figure 3A, the dia2Δ mrc1Δ mutant exhibited modestly stronger growth than the dia2Δ strain on media containing MMS.
Figure 3 .
Checkpoint-defective alleles of mrc1 suppress dia2Δ MMS sensitivity and checkpoint recovery defects. (A) mrc1 mutant alleles suppress dia2Δ MMS sensitivity. Tenfold serial dilutions of the indicated strains were spotted on YPD or YPD + 0.007% MMS and incubated at 30°. (B) Checkpoint-defective mrc1 alleles enhance viability of dia2Δ in MMS. Equal numbers of cells were plated on media containing the indicated amounts of MMS, and colony-forming units were counted after 4 days at 30°. Error bars represent standard deviations from three independent experiments. (C) mrc11–971 does not suppress pph3Δ MMS sensitivity. Tenfold serial dilutions of the indicated strains were spotted on YPD or YPD + 0.01% MMS and incubated at 30°. (D and E) Checkpoint-defective mrc1 alleles accelerate dia2Δ checkpoint recovery. Cells were arrested in late G1 by α-factor, (D) released into YPD + 0.033% MMS for 40 min, and then released into YPD or, (E) released into YPD at 30°. 1C and 2C indicate DNA content. (F) mrc11–971 and mrc1AQ accelerate Rad53 deactivation of dia2Δ. Cells were arrested in G1 by α-factor, released in YPD + 0.009% MMS for 1 hr, and then released into YPD + 15 μg/ml nocodazole. Protein samples were taken as indicated. Rad53-P and Rad53 represent phosphorylated and unphosphorylated Rad53 proteins, respectively. The very top modified band of Rad53 was quantified using ImageJ and the percentage of that in each time point relative to time zero is shown in the graph.
Since the suppressor mrc1 allele was checkpoint defective, we asked if the reduction of Mrc1-mediated checkpoint function was important for the suppression of dia2Δ cells. We tested whether mrc1AQ, a previously described checkpoint-defective allele in which all S/TQ phosphosites were mutated to AQ (Osborn and Elledge 2003), could also rescue the MMS sensitivity of the dia2Δ strain. Growth and viability of dia2Δ, dia2Δ mrc11–971, and dia2Δ mrc1AQ cells on MMS-containing media were assayed as previously described. We found that checkpoint-defective mrc11–971 and mrc1AQ mutants at least modestly enhanced growth and viability of dia2Δ cells in the presence of MMS (Figure 3, A and B). The suppression by mrc11–971 is specific to dia2Δ cells, as we did not observe mrc11–971 suppressing the MMS sensitivity of pph3Δ (Figure 3C). We then investigated whether the mrc11–971 and the mrc1AQ mutants would suppress the checkpoint recovery defect of dia2Δ cells. Cells were arrested in G1, released into MMS-containing media, and then released into media without MMS to observe DNA replication recovery when the checkpoint was deactivated (Figure 3D). Cells were also released from G1 into S-phase in media without MMS as a control (Figure 3E). As shown in Figure 3D, wild-type cells completed DNA replication by 60 min, whereas the majority of dia2Δ cells did not finish DNA replication until the last time point of the experiment. Similar to wild-type cells, the majority of dia2Δ mrc11–971 and dia2Δ mrc1AQ cells completed DNA replication by 60 min. Thus, the mrc11–971 and mrc1AQ mutants suppress the checkpoint recovery defect of the dia2Δ strain.
Previous work has shown that Rad53 is constitutively hyperphosphorylated in dia2Δ cells (Pan et al. 2006), but it has not been established whether this is a result of failure to deactivate the checkpoint or constant reinitiation of checkpoint signaling. If Dia2 is critical for checkpoint recovery, we would expect to see a defect in Rad53 deactivation in dia2Δ cells. To test this, cells were arrested in G1, released into MMS-containing media to activate Rad53, and then released into media containing nocodazole. Nocodazole was used to block the cell cycle at early G2/M to separate Rad53 deactivation during S-phase checkpoint recovery from G2/M checkpoint recovery. As shown in Figure 3F, the top phosphorylated Rad53 band was more intense in the dia2Δ strain compared to wild type at the 1.5-hr time point. Thus, Rad53 deactivation was slower in dia2Δ cells than in wild type.
Since the mrc11–971 and the mrc1AQ mutants suppress the checkpoint recovery defect of dia2Δ cells, we would expect Rad53 deactivation to be more robust in dia2Δ mrc11–971 and dia2Δ mrc1AQ cells because the checkpoint would not be fully activated in the first place, making recovery faster despite the lack of Dia2. Consistent with the mrc1 alleles being checkpoint defective, we found fewer phosphorylated Rad53 bands in the mrc11–971 and mrc1AQ strains compared to wild type at time zero of checkpoint recovery, and the same was true in the dia2Δ mrc11–971 and the dia2Δ mrc1AQ mutants compared to the dia2Δ strain. Not surprisingly, the top phosphorylated Rad53 band in dia2Δ mrc11–971 and dia2Δ mrc1AQ cells was less intense than that in dia2Δ cells at the 1.5-hr time point (Figure 3F). Indeed, the intensity of the top bands was similar between wild-type, dia2Δ mrc11–971, dia2Δ mrc1AQ cells. These data suggest that checkpoint recovery is faster in the dia2Δ mrc1 double mutants because the initial activation of the S-phase checkpoint is not as robust.
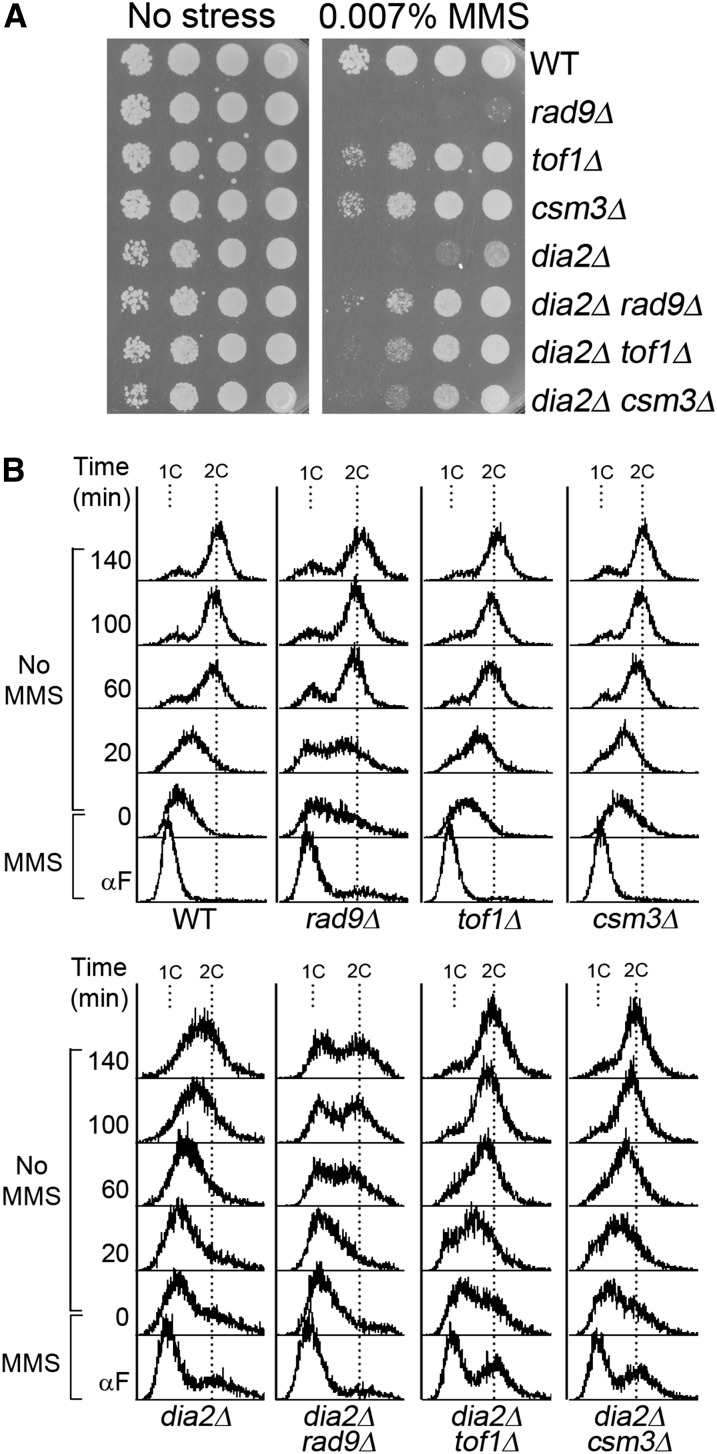
Since Rad9, Tof1, and Csm3 are mediators of Rad53 checkpoint phosphorylation in addition to Mrc1, we predict that rad9, tof1, and csm3 mutants would also suppress dia2Δ due to a lower level of Rad53 checkpoint activation. To test this, dia2Δ, dia2Δ rad9Δ, dia2Δ tof1Δ, and dia2Δ csm3Δ cells were analyzed for growth on MMS-containing media (Figure 4A) and checkpoint recovery by flow cytometry (Figure 4B). As expected, rad9Δ, tof1Δ, and csm3Δ mutants all, at least partially, suppress dia2Δ MMS sensitivity and checkpoint recovery defects (Figure 4, A and B). Interestingly, the dia2Δ and rad9Δ mutants appear to mutually suppress each other’s MMS sensitivity. The explanation for this phenotype is unclear, but we note that rad9Δ suppression of the dia2Δ recovery defect is the least robust among the Rad53 mediators, suggesting that Rad9 and Dia2 may function together in another aspect of the cellular response to MMS. Overall, our data suggest that removal of Rad53 mediators suppresses dia2Δ MMS sensitivity and recovery from an MMS-induced checkpoint.
Figure 4 .
Rad53 mediator mutants suppress dia2Δ MMS sensitivity and checkpoint recovery defects. (A) rad9Δ, tof1Δ, and csm3Δ mutants suppress dia2Δ MMS sensitivity. Tenfold serial dilutions of the indicated strains were spotted on YPD or YPD + 0.007% MMS and incubated at 30°. (B) rad9Δ, tof1Δ, and csm3Δ mutants accelerate dia2Δ checkpoint recovery. Cells were arrested in G1 by α-factor, released into YPD + 0.033% MMS for 40 min, and then released into YPD at 30°. 1C and 2C indicate DNA content.
Dia2 targets Mrc1 for degradation during checkpoint recovery
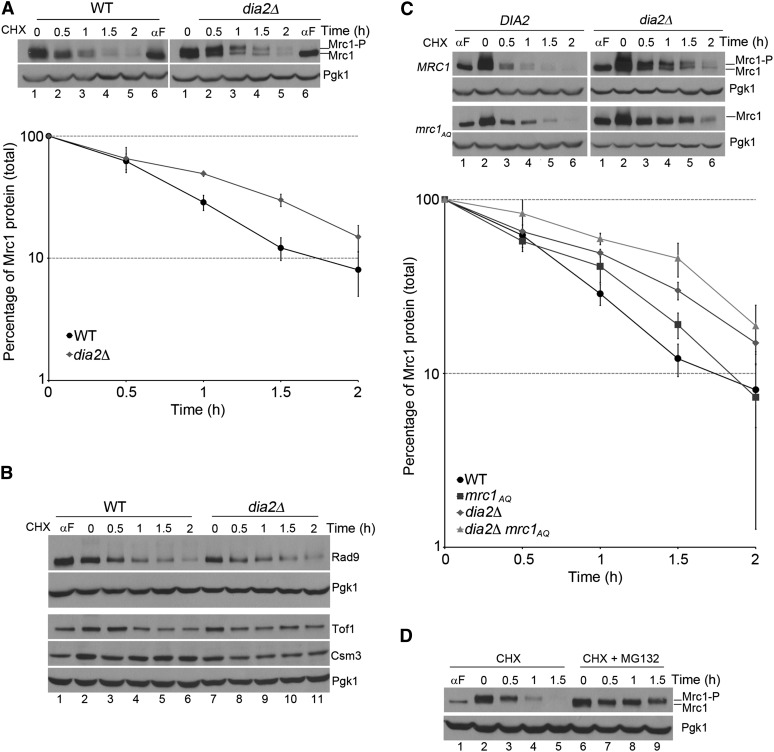
Our data raised the possibility that Dia2 mediates Mrc1 degradation for checkpoint recovery. If this were the case, we would predict that Dia2 targets Mrc1 for degradation to facilitate inactivation of checkpoint signaling and return to the cell cycle. To test this hypothesis, we monitored the stability of Mrc1 in wild-type and dia2Δ cells during S-phase checkpoint recovery. Cells were arrested in G1, released into MMS-containing media for S-phase checkpoint activation for 40 min, and then released into media containing cycloheximide (CHX) to stop protein synthesis during checkpoint inactivation. Protein samples were taken every half hour during CHX treatment to determine Mrc1 stability over time as cells progressed into the cell cycle. We found that the level of Mrc1 protein decreased during checkpoint recovery in wild-type cells. Interestingly, the Mrc1 protein was partially stabilized in dia2Δ cells (Figure 5A). It appears that both phosphorylated and unmodified Mrc1 are stabilized in dia2Δ cells. These results are consistent with the hypothesis that Dia2 targets Mrc1 for degradation to facilitate checkpoint recovery.
Figure 5 .
Dia2 is required for the degradation of Mrc1 during checkpoint recovery. (A) Mrc1 is degraded in a Dia2-dependent manner. Cells were arrested in G1 by α-factor, released into YPD + 0.033% MMS for 40 min, and then released into YPD + 200 μg/ml CHX. Protein samples were taken at indicated times. Mrc1-P and Mrc1 represent phosphorylated and unphosphorylated Mrc1 proteins, respectively. Pgk1 serves as a loading control. The graph shows the quantification of three independent experiments. Error bars indicate standard deviations. (B) Rad9, Csm3, and Tof1 are not degraded in a Dia2-dependent manner during recovery from an MMS-induced checkpoint. The experiment was performed as in A. Pgk1 serves as a loading control. (C) S/TQ phosphosites play a role in the degradation of Mrc1 during checkpoint recovery. Cells were treated as described in A. Mrc1-P and Mrc1 represent phosphorylated and unphosphorylated Mrc1 proteins, respectively. Pgk1 serves as a loading control. Stability of total Mrc1 protein was quantified from three independent experiments. Error bars were derived from standard deviations of the three experiments. (D) Degradation of phosphorylated Mrc1 is proteasome dependent. Wild-type cells were subjected to the same arrest and release treatment as described in A, except that during checkpoint recovery one set of cells was released into YPD + 200 μg/ml CHX and another set into YPD + 200 μg/ml CHX + 50 μM MG-132. Mrc1-P and Mrc1 represent phosphorylated and unphosphorylated Mrc1 proteins, respectively. Pgk1 serves as a loading control.
Because rad9Δ, tof1Δ, and csm3Δ mutants also suppress dia2Δ MMS sensitivity and checkpoint recovery defects, we tested if these Rad53 mediators are also targeted for degradation during checkpoint recovery. Although Rad9 appeared to be slowly degraded during recovery, the turnover rate was indistinguishable for Rad9, Tof1, and Csm3 proteins in wild-type and dia2Δ cells (Figure 5B). These data suggest that Dia2 specifically targets Mrc1 among Rad53 mediators for degradation during checkpoint recovery.
Next, we investigated the mechanism of Dia2 targeting Mrc1 for degradation to facilitate checkpoint inactivation. We asked if Dia2 targets checkpoint-activated Mrc1 for degradation by recognizing specific sites on Mrc1 critical for its checkpoint function. One well-characterized mechanism of Mrc1 checkpoint activation is phosphorylation of S/TQ sites by Mec1 (Osborn and Elledge 2003). We tested if the S/TQ phosphosites are essential for Dia2-mediated Mrc1 degradation during checkpoint recovery. Wild-type, mrc1AQ, dia2Δ, and dia2Δ mrc1AQ cells were treated as described for Figure 5A. The amount of total Mrc1 protein was measured over time. We observed that the Mrc1AQ protein is partially stabilized compared to wild-type Mrc1 (Figure 5C), suggesting that S/TQ phosphorylation contributes to Mrc1 degradation during checkpoint recovery. However, the extent of stabilization exhibited by the Mrc1AQ protein is not as robust as the stabilization of the wild-type Mrc1 protein in the absence of DIA2, indicating that S/TQ phosphorylation is not the only determinant of Mrc1 degradation. In addition, the stabilization of the Mrc1AQ protein is slightly enhanced in dia2Δ cells (Figure 5C), suggesting that another mechanism in addition to Dia2 may mediate Mrc1 degradation during checkpoint recovery. Nevertheless, the results are consistent with a model in which Dia2 mediates degradation of Mrc1 during recovery from MMS-induced DNA damage.
If the change in the stability of Mrc1 was due to ubiquitin-mediated degradation, we would expect that the stability would be proteasome dependent. To test this, wild-type cells were subjected to the same treatment as described for Figure 5A, except that during checkpoint recovery one set of cells was released into CHX-containing media and the other set was released into media containing both CHX and the proteasome inhibitor MG-132. As shown in Figure 5D, checkpoint-activated Mrc1 was strongly stabilized in the presence of MG-132. These data suggest that phosphorylated Mrc1 is targeted by Dia2 for proteasome-dependent degradation during checkpoint recovery.
A previous study identified two putative Dia2 binding regions in Mrc1 by yeast two-hybrid analysis using mrc1 fragments (Mimura et al. 2009). These two regions span residues 380–557 and 701–800 of Mrc1 (Mimura et al. 2009) (supporting information, Figure S1A). We constructed two deletion mutants with one lacking residues 380–430 and 701–800 and another lacking residues 461–557 and 701–800. In an attempt to avoid the complication of compromising Mrc1 checkpoint function, we did not delete the region between residues 431 and 460 because that region was reported to be important for Rad53 checkpoint activation (Naylor et al. 2009) and smaller variations of the 380–557 region were shown to be insufficient for the two-hybrid interaction between Dia2 and Mrc1 (Mimura et al. 2009). If these putative Dia2-binding regions are important for degradation during recovery, we would expect that deleting these regions would stabilize Mrc1 proteins relative to full-length Mrc1 and that no difference in stability would be observed in dia2Δ cells. However, the deletion of these two-hybrid Dia2-binding regions in Mrc1 do not stabilize the protein in a wild-type background and these mutant proteins are still stabilized in dia2Δ cells (Figure S1B). Consistent with the stability data, we did not observe any checkpoint recovery defect in these mrc1 mutants (Figure S1C). These data indicate that these domains are not required for Dia2-dependent turnover of Mrc1.
S. cerevisiae Mrc1 has two overlapping DSGxxS sequences that are analogous to residues important for Claspin degradation during G2/M recovery in humans (Mamely et al. 2006; Peschiaroli et al. 2006) (Figure S1D). We mutated the three serine residues within the sequence to alanine (mrc13SA) and examined the mutant for Mrc1 stability and checkpoint recovery. We found that the stability of Mrc1 was indistinguishable between wild-type and mrc13SA cells (Figure S1D). Furthermore, wild-type and mrc13SA cells completed DNA replication at the same rate during checkpoint recovery (Figure S1E). These data suggest that the DSGxxS motifs are not required for S-phase checkpoint recovery or the degradation of Mrc1 during recovery and that Mrc1 degradation in S-phase is unlikely to be the analogous pathway to Claspin degradation in G2/M recovery.
Degradation of checkpoint-functional Mrc1 contributes to Dia2-mediated S-phase checkpoint recovery
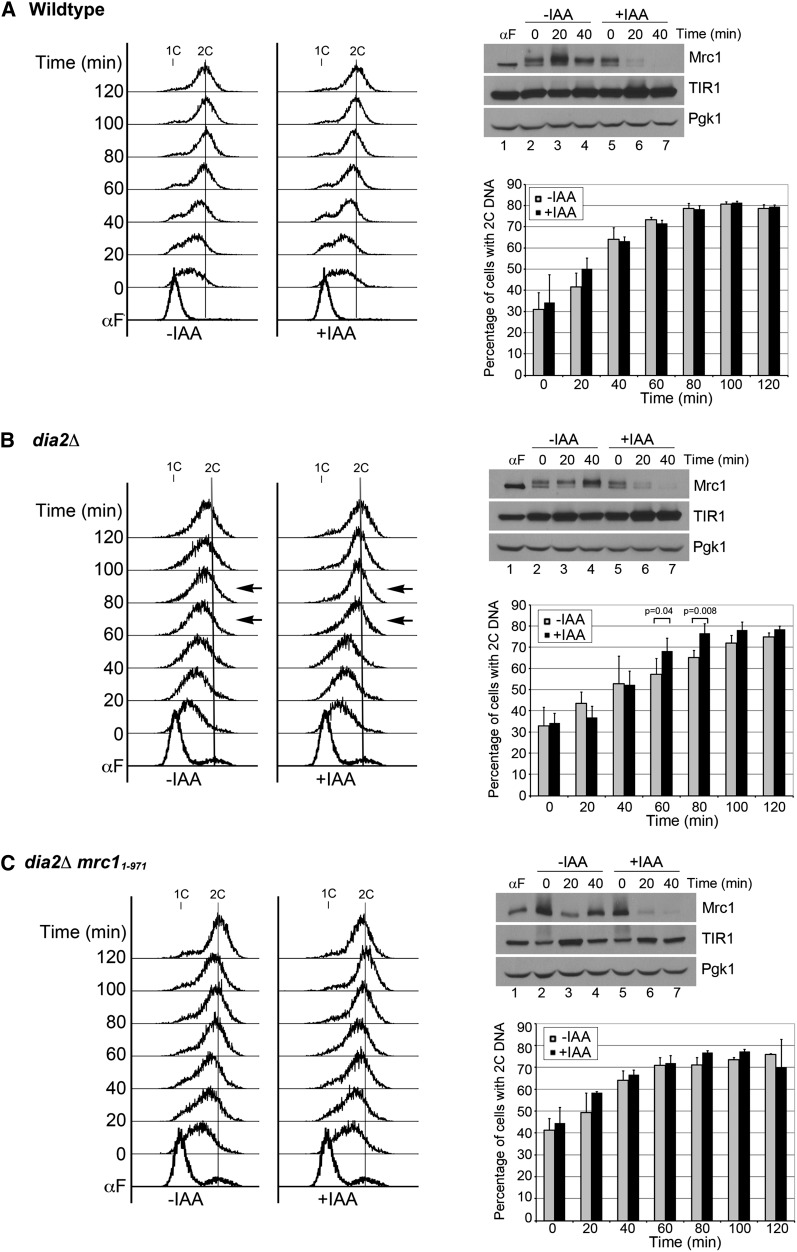
One explanation for our results is that Dia2 mediates Mrc1 degradation to help facilitate cell-cycle reentry during checkpoint recovery. If this were the case, we would expect the degradation of Mrc1 to be important for the resumption of DNA replication in the dia2Δ strain after cells have been exposed to MMS. To test this directly, we utilized the auxin-inducible degron system (Dreher et al. 2006; Nishimura et al. 2009) to induce degradation of Mrc1 in wild-type and dia2Δ cells during S-phase checkpoint recovery. Cells were arrested in G1, released into media containing MMS for checkpoint activation, and then released into media without MMS to observe DNA replication recovery when the checkpoint was deactivated. During the time course in media without MMS, one set of cells was treated with auxin IAA to induce degradation of Mrc1 whereas another set of cells did not receive IAA. To verify that the auxin-induced degradation functioned appropriately, protein samples were taken 20 and 40 min after cells were released from the MMS-induced checkpoint to observe Mrc1 protein levels. In all strains, Mrc1 was rapidly degraded in the presence of IAA and was significantly reduced after 20 min and barely detectable after 40 min of treatment (Figure 6, A–C, top right). Cell-cycle progression was monitored by flow cytometry (Figure 6, A–C, left) and the percentage of cells reaching 2C DNA content was quantified from multiple experiments (Figure 6, A–C, bottom right). In the wild-type control strain, 70% or more cells had achieved 2C DNA content by 60 min regardless whether the cells were treated with IAA. Induced degradation of Mrc1 did not significantly change the kinetics of DNA replication recovery in wild-type cells post-MMS exposure. This result also suggests that ongoing degradation of Mrc1 does not noticeably slow down DNA replication during checkpoint recovery from MMS-induced DNA damage.
Figure 6 .
Induced degradation of checkpoint-functional Mrc1 contributes to Dia2-mediated checkpoint recovery. Cells were arrested in G1 by α-factor, released into YPD + 0.033% MMS for 1 hr, and then released into either YPD + ethanol (vehicle) or YPD + 1.5 mM IAA (auxin) at 30°. Results for wild type are shown in A and dia2Δ in B. (C) The Mrc11–971 protein was degraded in a dia2Δ strain. Left: Cell-cycle progression was monitored by flow cytometry. 1C and 2C indicate DNA content. Arrows mark significant difference between –IAA and +IAA samples in the dia2Δ strain. Top right: Mrc1 was rapidly degraded upon IAA treatment. Protein samples were taken at indicated times with or without IAA treatment. Pgk1 serves as a loading control. Bottom right: Quantification of 2C DNA content from at least three replicates of each experiment. Error bars indicate standard deviations. P-values calculated using paired Student’s t-test analysis (n = 4).
When we compared the DNA replication profiles of untreated and IAA-treated dia2Δ cells, we observed that 70% of the IAA-treated cells reached 2C DNA content between the 60- and 80-min time points, whereas the untreated cells did not reach 70% 2C DNA content until much later in the time course. At both the 60- and 80-min time points, the percentage of IAA-treated dia2Δ cells that had completed DNA replication was significantly higher than untreated dia2Δ cells at these time points (Figure 6B, see arrows, left, and P-values on graph). Thus, induced degradation of Mrc1 accelerated the rate at which dia2Δ cells recovered from an MMS-induced checkpoint.
To test whether the acceleration of checkpoint recovery kinetics in dia2Δ cells is due to a downregulation of Mrc1 checkpoint signaling or a modulation of its replication function, we repeated the same experiment to induce degradation of checkpoint-defective but replication-proficient Mrc11–971 protein in dia2Δ cells (Figure 6C). Consistent with our earlier results, mrc11–971 suppressed dia2Δ checkpoint recovery defects. By 60 min, the difference between IAA-treated and -untreated cells was indistinguishable, as 70% of both untreated and treated dia2Δ mrc11–971 cells reached 2C DNA content. However, there was a slight increase in the percentage of cells with 2C DNA content in the IAA-treated samples at the 80- and 100-min time points. These data suggest that degradation of checkpoint-activated Mrc1 is more important in deactivation of checkpoint signaling than Mrc1 DNA replication activity. Altogether these results indicate that degradation of checkpoint-functional Mrc1 contributes to Dia2-mediated checkpoint recovery.
Discussion
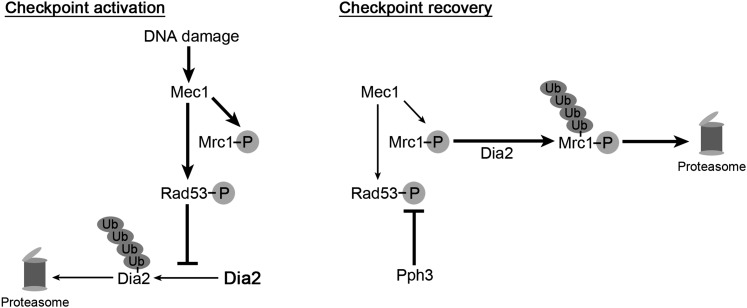
In this study, we identified Dia2 as a novel player in the S-phase checkpoint recovery network. By genetic analysis, Dia2 acts in a parallel pathway to Rad53 phosphatase Pph3. In principle, there are a number of possible roles for Dia2 in checkpoint recovery, such as repair of DNA damage or mechanisms that stimulate initiation of DNA replication. However, our results demonstrate that at least one mechanism involves degradation of the Rad53 mediator Mrc1. The Mrc1 protein is stabilized during checkpoint recovery in dia2Δ cells. Importantly, induced degradation of checkpoint-functional Mrc1 contributes to Dia2-mediated checkpoint recovery, whereas degradation of checkpoint-defective Mrc11–971 did not dramatically affect the kinetics of recovery. We favor a model in which Dia2 mediates the degradation of Mrc1 to promote the resumption of DNA replication during recovery from MMS-induced DNA damage in S-phase (Figure 7).
Figure 7 .
Model for the role of Dia2 in S-phase checkpoint recovery. (Left; Checkpoint activation) Dia2 is stabilized by the activation of the S-phase checkpoint (Kile and Koepp 2010). (Right; Checkpoint recovery) During checkpoint recovery, Dia2 targets checkpoint-activated Mrc1 for degradation to downregulate the checkpoint activation of Rad53. Rad53 phosphatase Pph3 removes phosphate groups from Rad53 to allow DNA replication to resume in S-phase (O’Neill et al. 2007; Szyjka et al. 2008). Ub, ubiquitin; P, phosphorylation.
It is a bit puzzling that Mrc1 is degraded during S-phase checkpoint recovery as Mrc1 has been shown to be required for normal replication efficiency (Szyjka et al. 2005) and stabilization of the replisome complex (Katou et al. 2003). Our results address only degradation of Mrc1 when cells resume the cell cycle during checkpoint recovery from an MMS-induced checkpoint. It is possible that the initial association of Mrc1 to the replisome complex allows stabilization of the replisome and efficient DNA replication. During checkpoint recovery, perhaps Mrc1 is degraded to decrease the checkpoint signal below a particular threshold without compromising the integrity of the replisome. Such a scenario may explain our observation that the auxin-induced degradation of Mrc1 did not noticeably affect the replication kinetics in wild-type cells during recovery from an MMS-induced checkpoint.
It is not clear whether Dia2 targets Mrc1 for degradation during recovery from activation of the S-phase checkpoint via other mechanisms that cause fork stalling, such as exposure to HU. Mimura et al. (2009) demonstrated that Mrc1, particularly the chromatin-associated population, was stabilized in dia2Δ cells when cells are arrested with HU, which could reflect either a checkpoint response or an S-phase-specific pathway. The turnover of Mrc1 during recovery from an HU block has not been explicitly examined, but there is evidence to suggest that Dia2 may not play as important a role in recovery from HU. The dia2Δ strain is only mildly hypersensitive to HU, whereas the MMS hypersensitivity is quite strong, suggesting that there is a limited requirement for Dia2 during the cellular response to hydroxyurea. In addition, the mrc11–971 mutant does not rescue the HU sensitivity of the dia2Δ strain (C. M. Fong and D. M. Koepp, unpublished observations). Such a discrepancy in cellular response between two drugs that initiate the checkpoint is not unprecedented, as Pph3 and Ptc2 are required for recovery from an MMS-induced checkpoint but not an HU-induced checkpoint (Travesa et al. 2008). It is possible that a Dia2-independent degradation pathway is more prominent during recovery from an HU arrest.
Our results suggest that Dia2 and Pph3 may act in parallel pathways during recovery from an MMS-induced checkpoint. The two pathways may work together for a common goal; Pph3 removes phosphates from checkpoint-activated Rad53, while Mrc1 degradation prevents Rad53 from further activation. It is interesting that checkpoint-activated Mrc1 is degraded rather than dephosphorylated like Rad53 during checkpoint recovery. However, the human homolog of Mrc1, Claspin, is also degraded for checkpoint inactivation, albeit during G2/M recovery (Mailand et al. 2006; Peschiaroli et al. 2006). Rad53 activation is also dependent on other checkpoint mediators such as Rad9, Tof1, Csm3 (Navas et al. 1996; Sun et al. 1998; Vialard et al. 1998; Foss 2001; Schwartz et al. 2002). We did not find mediators other than Mrc1 to be targeted by Dia2 for degradation during S-phase checkpoint recovery. As Mrc1 forms a complex with Csm3 and Tof1, there are clearly questions that remain to be answered in how Mrc1 is specifically targeted for degradation while Csm3 and Tof1 are spared and whether Csm3 or Tof1 are inhibited via other mechanisms. Intriguingly, Rad9 is also degraded during recovery from MMS, although the degradation is independent of Dia2. We speculate that diversity in the regulation of checkpoint recovery may be advantageous for cells in the event that one of these mechanisms fails.
In pph3Δ cells, replication forks fail to restart post-MMS exposure. Instead, cells appear to rely on late-firing origins to finish DNA replication passively (O’Neill et al. 2007; Szyjka et al. 2008). We speculate that similar defects may contribute to the slow DNA replication in dia2Δ cells during checkpoint recovery. However, when checkpoint-defective mrc1 alleles suppress the dia2Δ recovery defect, it may be because they allow more robust fork restart in dia2Δ cells, as Rad53 deactivation is faster in these cells. This model is consistent with the report showing that Rad53 deactivation is sufficient for fork restart in pph3Δ cells during recovery (Szyjka et al. 2008). Alternatively, late-firing origins may initiate prematurely in dia2Δ mrc11–971 and dia2Δ mrc1AQ cells, allowing the faster completion of DNA replication post-MMS exposure. This alternative is consistent with data showing that Rad53 checkpoint activation is critical to protecting origins from firing inappropriately in the presence of MMS (Santocanale and Diffley 1998; Shirahige et al. 1998; Tercero and Diffley 2001). Future work will be needed to distinguish between these possibilities.
Our results suggest that recognition of Mrc1 by Dia2 is complex, as both checkpoint-phosphorylated and unmodified Mrc1 are stabilized in dia2Δ cells. However, Mrc1AQ is only partially stabilized relative to wild-type Mrc1 and its stability is slightly enhanced in dia2Δ cells. One possible explanation is that a change of Mrc1 conformation and perhaps protein–protein interaction upon S/TQ phosphorylation triggers the degradation of phosphorylated Mrc1 protein during checkpoint recovery. This possibility is consistent with a previously proposed model in which S/TQ phosphorylation of Mrc1 changes its conformation and association with replisome components (Lou et al. 2008). It is possible that Dia2 maintains a basal level of association with Mrc1 in S-phase and their interaction is strengthened by the phosphorylation-dependent conformational change to Mrc1, leading to enhanced Mrc1 degradation during checkpoint recovery. Alternatively, it is possible that Dia2 recognizes one or more of the identified domains (Naylor et al. 2009) that lack S/TQ and yet are required for checkpoint activation to trigger Mrc1 degradation. That the Mrc1AQ protein is modestly stabilized in dia2Δ cells is consistent with an additional S/TQ-dependent mechanism for Mrc1 degradation during checkpoint recovery. It will be interesting to elucidate the regulation of Mrc1 degradation in future studies.
A previous study identified two putative Dia2-interacting regions by yeast two-hybrid analysis with mrc1 fragments of various lengths (Mimura et al. 2009). However, the study used only fragments of Mrc1 in a directed two-hybrid test, so the importance of each domain for Mrc1 protein integrity and function was not determined. Regardless, the two regions are unlikely to contain the Dia2-specific degron because deletion of the regions did not stabilize Mrc1. When we mutated the DSGxxS phosphosites similar to those important for Claspin degradation during G2/M checkpoint recovery (Mamely et al. 2006; Peschiaroli et al. 2006), we did not see any defect in S-phase checkpoint recovery or stabilization of Mrc1. Our findings raise the question of whether Claspin degradation is also regulated for S-phase checkpoint recovery in mammalian cells.
Our genetic screen identified a new checkpoint-defective allele of MRC1 that is truncated at the C terminus but retains all of the S/TQ phosphosites. This came as a surprise because previous studies reported that these phosphosites of Mrc1 are critical for checkpoint activation whereas the C terminus is important for its replication function (Osborn and Elledge 2003; Naylor et al. 2009). We speculate that the contradiction is perhaps due to a difference in the methods used to assay for checkpoint activation, as impaired checkpoint function of the mrc11–971 allele was more obvious when other checkpoint proteins were also defective. Our results also show that the Mrc11–971 protein is not efficiently hyperphosphorylated during checkpoint activation, which may contribute to its checkpoint defect. Our findings suggest that the structure and function relationship of Mrc1 is more complex than originally thought.
Overall, this work expands our knowledge on how cells recover from genotoxin-induced checkpoints. A better understanding of checkpoint recovery may lead to more efficient cancer treatments. The S-phase checkpoint is a target of interest for antitumor therapies because chemotherapy and radiotherapy induce genotoxic stress to trigger cell death in cancer cells. Inhibitors of checkpoint activators are currently being explored as treatments to overload cancer cells with genotoxic stress (Chen et al. 2011). Thus, checkpoint recovery components may serve as alternative targets of treatment to sensitize cancer cells to antitumor therapies.
Supplementary Material
Acknowledgments
We thank Stephen J. Elledge (Harvard Medical School) for strains and reagents. We thank Dong-Hwan Kim, Owen Smith, Alisha Bailey, Allison Bock, and Matthew Schwartz for technical assistance. This work was funded by the National Institutes of Health grant R01GM076663.
Footnotes
Communicating editor: O. Cohen-Fix
Literature Cited
- Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J., et al. , 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965 [DOI] [PubMed] [Google Scholar]
- Ang X. L., Harper J.W., 2005. SCF-mediated protein degradation and cell cycle control. Oncogene 24: 2860–2870 [DOI] [PubMed] [Google Scholar]
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., et al. , 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas J., 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19: 238–245 [DOI] [PubMed] [Google Scholar]
- Blake D., Luke B., Kanellis P., Jorgensen P., Goh T., et al. , 2006. The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics 174: 1709–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M., 2005. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17: 568–575 [DOI] [PubMed] [Google Scholar]
- Chen S. H., Zhou H., 2009. Reconstitution of Rad53 activation by Mec1 through adaptor protein Mrc1. J. Biol. Chem. 284: 18593–18604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Stephens P. A., Middleton F. K., Curtin N. J., 2011. Targeting the S and G2 checkpoint to treat cancer. Drug Discov. Today 17: 194–202 [DOI] [PubMed] [Google Scholar]
- Cimprich K. A., Cortez D., 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V., Shechter D., Lupardus P. J., Cimprich K. A., Gottesman M., et al. , 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11: 203–213 [DOI] [PubMed] [Google Scholar]
- Deshaies R. J., 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15: 435–467 [DOI] [PubMed] [Google Scholar]
- Dreher K. A., Brown J., Saw R. E., Callis J., 2006. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274: 1664–1672 [DOI] [PubMed] [Google Scholar]
- Fanning E., Klimovich V., Nager A. R., 2006. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34: 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman R. M., Correll C. C., Kaplan K. B., Deshaies R. J., 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91: 221–230 [DOI] [PubMed] [Google Scholar]
- Foss E. J., 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157: 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C. S., Green C. M., Lowndes N. F., 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8: 129–136 [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A., 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246: 629–634 [DOI] [PubMed] [Google Scholar]
- Kamura T., Koepp D. M., Conrad M. N., Skowyra D., Moreland R. J., et al. , 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284: 657–661 [DOI] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., et al. , 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Kile A. C., Koepp D. M., 2010. Activation of the S-phase checkpoint inhibits degradation of the F-box protein Dia2. Mol. Cell. Biol. 30: 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp D. M., Kile A. C., Swaminathan S., Rodriguez-Rivera V., 2006. The F-box protein Dia2 regulates DNA replication. Mol. Biol. Cell 17: 1540–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Wakayama T., Naiki T., Matsumoto K., Sugimoto K., 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294: 867–870 [DOI] [PubMed] [Google Scholar]
- Lou H., Komata M., Katou Y., Guan Z., Reis C. C., et al. , 2008. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol. Cell 32: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Bartek J., Lukas J., 2006. Destruction of claspin by SCF beta TrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 23: 307–318 [DOI] [PubMed] [Google Scholar]
- Mamely I., Van Vugt M. A. T. M., Smits V. A. J., Semple J. I., Lemmens B., et al. , 2006. Polo-like kinase-1 controls proteasome-dependent degradation of claspin during checkpoint recovery. Curr. Biol. 16: 1950–1955 [DOI] [PubMed] [Google Scholar]
- Melo J. A., Cohen J., Toczyski D. P., 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15: 2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S., Komata M., Kishi T., Shirahige K., Kamura T., 2009. SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. EMBO J. 28: 3693–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas T. A., Sanchez Y., Elledge S. J., 1996. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 10: 2632–2643 [DOI] [PubMed] [Google Scholar]
- Naylor M. L., Li J. M., Osborn A. J., Elledge S. J., 2009. Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. Proc. Natl. Acad. Sci. USA 106: 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M., 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6: 917–922 [DOI] [PubMed] [Google Scholar]
- O’Connell M. J., Walworth N. C., Carr A. M., 2000. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 10: 296–303 [DOI] [PubMed] [Google Scholar]
- O’Neill B. M., Szyjka S. J., Lis E. T., Bailey A. O., Yates J. R., et al. , 2007. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. USA 104: 9290–9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn A. J., Elledge S. J., 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., et al. , 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., et al. , 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18: 6561–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschiaroli A., Dorrello N. V., Guardavaccaro D., Venere M., Halazonetis T., et al. , 2006. SCF beta TrCP-mediated degradation of claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 23: 319–329 [DOI] [PubMed] [Google Scholar]
- Rhind N., Russell P., 2000. Checkpoints: it takes more than time to heal some wounds. Curr. Biol. 10: R908–R911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. (), 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Santocanale C., Diffley J. F., 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Schwartz M. F., Duong J. K., Sun Z., Morrow J. S., Pradhan D., et al. , 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9: 1055–1065 [DOI] [PubMed] [Google Scholar]
- Shimada K., Oma Y., Schleker T., Kugou K., Ohta K., et al. , 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 18: 566–575 [DOI] [PubMed] [Google Scholar]
- Shirahige K., Hori Y., Shiraishi K., Yamashita M., Takahashi K., et al. , 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W., 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219 [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Lopes M., Foiani M., 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Sun Z., Hsiao J., Fay D. S., Stern D. F., 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281: 272–274 [DOI] [PubMed] [Google Scholar]
- Sweeney F. D., Yang F., Chi A., Shabanowitz J., Hunt D. F., et al. , 2005. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr. Biol. 15: 1364–1375 [DOI] [PubMed] [Google Scholar]
- Szyjka S. J., Viggiani C. J., Aparicio O. M., 2005. Mrc1 is required for normal progression of replication forks throughout chromatin in S-cerevisiae. Mol. Cell 19: 691–697 [DOI] [PubMed] [Google Scholar]
- Szyjka S. J., Aparicio J. G., Viggiani C. J., Knott S., Xu W., et al. , 2008. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 22: 1906–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J. A., Diffley J. F., 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., et al. , 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- Travesa A., Duch A., Quintana D. G., 2008. Distinct phosphatases mediate the deactivation of the DNA damage checkpoint kinase Rad53. J. Biol. Chem. 283: 17123–17130 [DOI] [PubMed] [Google Scholar]
- Van Vugt M. A., Medema R. H., 2004. Checkpoint adaptation and recovery: back with Polo after the break. Cell Cycle 3: 1383–1386 [DOI] [PubMed] [Google Scholar]
- Vialard J. E., Gilbert C. S., Green C. M., Lowndes N. F., 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17: 5679–5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H., 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241: 317–322 [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S. J., 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.