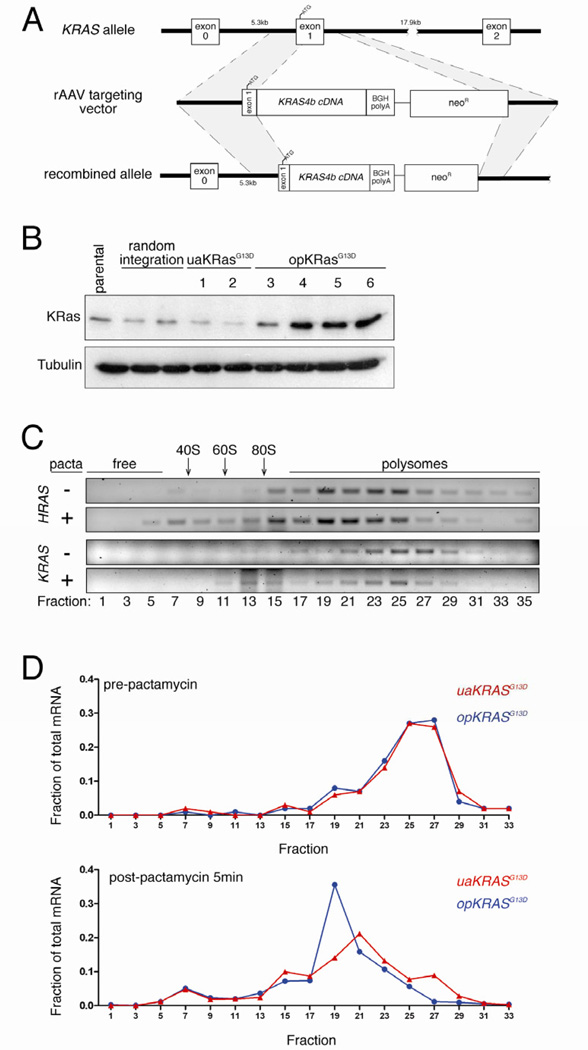
Figure 2. Rare codon bias limits endogenous KRAS translation.
(A) Targeting strategy to knock into exon 1 the endogenous KRAS gene in human HCT116 colon cancer cells (ATCC) oncogenic KRASG13D cDNA in which 130 rare codons were either optimized to common codons (opKRASG13D) or left unaltered (uaKRASG13D) by AAV-based recombination [16]. (B) Immunoblot of lysates isolated from parental HCT116 cell line, stable clones with non-homologous vector integration, clones with homologous integration of uaKRASG13D, and clones with homologous integration of opKRASG13D, with an αKRas or αtubulin antibody. One of one to two experiments. (C) Semi-quantitative RT-PCR using primers specific for HRAS and the non-targeted KRAS mRNA of the indicated sedimentation fractions (relative positions of ribosome-free, ribosome subunits 40S, 60S and 80S and polysome fractions) isolated from HCT116 opKRASG13D clone 5 pre- (-) and post- (+) pactamycin (pacta) treatment to halt translation initiation. (D) Quantitative RT-PCR using primers specific for the KRASG13D knock-in mRNA of the indicated sedimentation fractions isolated from HCT116 opKRASG13D clone 5 and uaKRASG13D clone 1 pre- and post-pactamycin treatment to halt translation initiation. Detailed methodologies and regent descriptions are provided in Supplemental Experimental Procedures. See also Figure S2.