Table II. Comparison between proteins identified by MALDI TOF MS from 2D-DIGE experiments including 44 patients and from LC-MS/MS validation experiments including 21 patients (a subgroup of patients included in the 2D-DIGE experiments). Significant proteins according to multivariate statistics indicated. aDark green and blue indicate statistical behavior of the majority of protein spots in a particular protein cluster, while light blue and green on the other hand indicate the behavior of at least one of the spots for a particular protein. In the cases where all protein spots are not-significant, the color is white. Dilated (D) and nondilated (C) thoracic aortic samples were examined. See Supplemental Table S4 for identification details on the LC-MS/MS analysis.
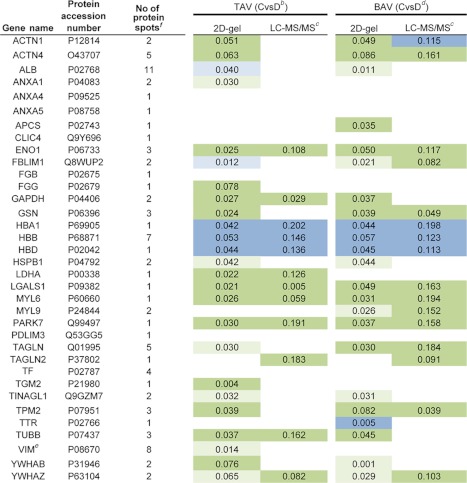
a The statistical analyses are based on non dilated versus dilated thoracic aorta samples in TAV and BAV (2D gel results are derived from Table I, with eight proteins not found in LC-MS/MS validation excluded).
b ABS (loading)-ABS(jack-knife conf interval) protein expression data, dilated higher (green), nondilated higher (blue), TAV.
c LC-MS/MS validation of 21 patients (a subgroup of patients included in the original 2D-DIGE analysis).
d ABS (loading)-ABS(jack-knife conf intervall) protein expression data, dilated higher (green), nondilated higher (blue), BAV.
e Previously published.
f Number of protein spots identified on the gel in the 2D-DIGE experiments.
