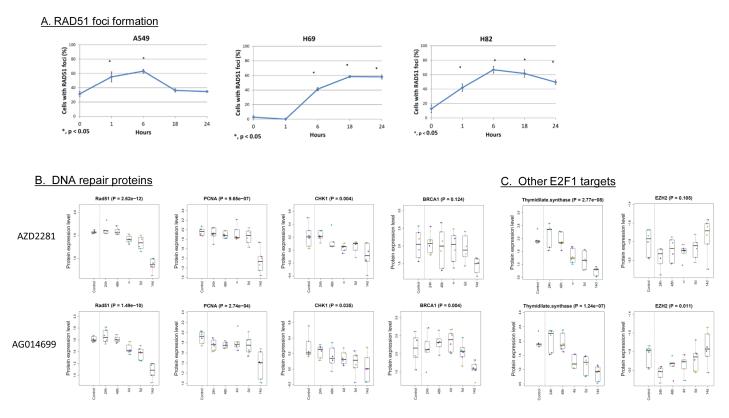
Figure 5. RAD51 foci formation (A) and modulation of proteins levels after PARP inhibitor treatment (B).
(A) Protein is localized at DNA DSBs region in response to stalled or collapsed DNA replication forks in SCLC (H69 & H82) but not in NSCLC cell (A549). Kinetics of RAD51 focus formation in NSCLC A549, SCLC H69, and SCLC H82. The percentage of cells with more than 5 nuclear foci was calculated. In each experiment, 100 nuclei were counted per data point. Error bars indicate standard error compared to unirradiated samples (*p<0.05). (B-C) Protein lysate was collected from three SCLC cell lines (H69, H82, H841) in duplicate at multiple timepoints (0-14d) after treatment with the PARP inhibitors AZD2281 and AGO14699. A time-dependent decrease was observed in multiple DNA repair proteins (B) and in other E2F1 targets such as thymidylate synthase (TS) and EZH2 (C). Note that TS follows a similar pattern to the other DNA repair proteins, while EZH2 is suppressed at 24h but recovers to baseline levels by 14d.