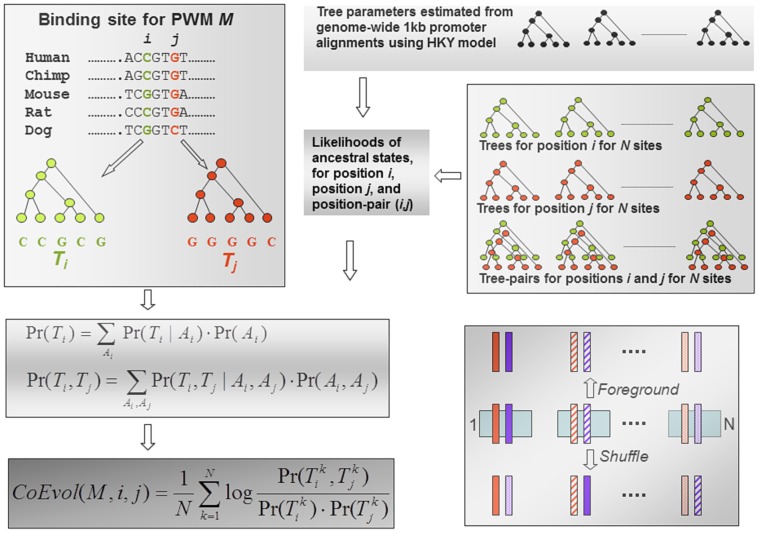
Figure 1. Overview of the approach.
The top left panel depicts positions i and j within a binding site for a PWM M having N genome-wide matches. The binding site in human is shown in the context of a 5-species multiple alignment. The top left panel also shows the phylogenetic trees for the two positions. The phylogenetic parameters are estimated from the genome-wide set of promoter alignments (top right panel). The likelihoods of ancestral nodes for a specific PWM and position (or position-pair) are then estimated from the N instances and the phylogenetic parameters. The likelihoods of individual trees and tree-pairs and ultimately the CoEvol for a pair of PWM positions are then estimated as detailed in the text. The lower right panel illustrates the procedure to generate Shuffle control. The figure depicts N instances of position pair (i,j) in the central row. A random j-position is paired with each of the i-positions (lower row).