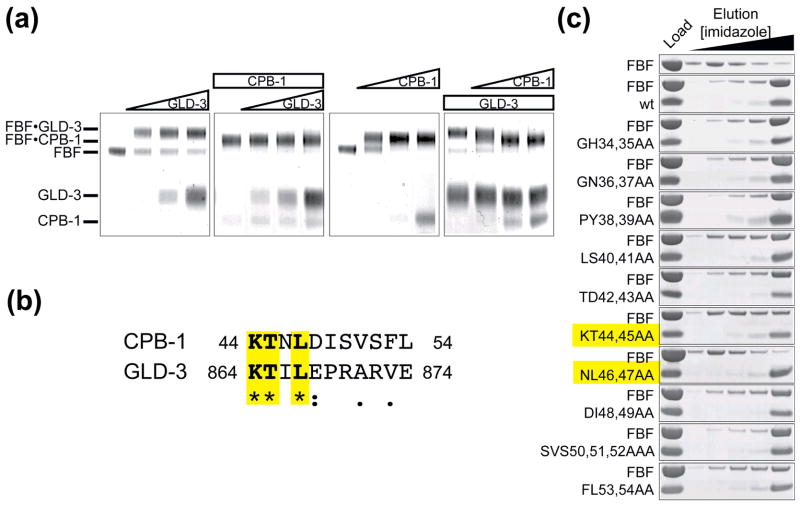
Figure 2. Sequence alignment with GLD-3 shows sequence conservation between the FBF-binding region of GLD-3 and CPB-1. CPB-1 and GLD-3 do not co-exist on FBF.
(A) CPB-1 and GLD-3 do not form a ternary complex with FBF. Protein complexes were covalently linked before analysis by SDS PAGE. The in-gel mobility of proteins alone and in complex is indicated on the left. The first panel from the left shows GLD-3 titration into a constant amount of FBF (increasing GLD-3 concentration is indicated with a triangle above the gel). The second panel shows GLD-3 titration to FBF pre-incubated with CPB-1 (constant amounts of protein are indicated with a rectangle). In the last two panels the order of addition of CPB-1 and GLD-3 to FBF was reversed. Increasing amounts of CPB-1 were added to FBF (third panel) and finally increasing amounts of CPB-1 were added to FBF pre-incubated with GLD-3. In the last experiment CPB-1 replaced GLD-3 on FBF. (B) Sequence alignment of CPB-1 with the FBF-binding region of GLD-3 (residues 864–874). Conserved residues are highlighted. The alignment was performed with ClustalW46, asterisks indicate positions that have a fully conserved residue, colons indicates conservation between groups of strongly similar properties and periods indicates conservation between groups of weakly similar properties. (C) Pulldown assay with FBF and CPB-1 double alanine mutants. Residues that most affected binding to FBF were located in the conserved 40–47 region.