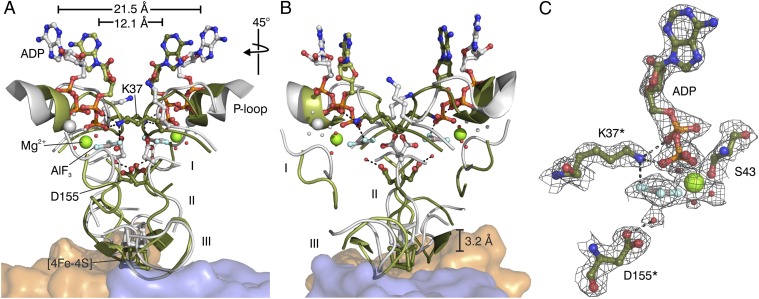
Fig. 2.
Dynamic switch mechanism of DPOR. (A) Superposition of the “on state” conformation of L2 in the ADP•AlF3–stablized complex (green) and the “off state” conformation of L2 in the ADP-bound state (gray, PDB ID code 3FWY). Nucleotide-dependent conformational rearrangements trigger the affinity of L2 for the (NB)2 core (orange and blue). The [4Fe–4S] cluster of L2 moves ∼3 Å toward the [4Fe–4S] cluster of (NB)2. Peptide segments undergoing large Cα-rearrangements are marked I (Asp66–Asp70 of switch I), II (Leu154–Cys158 of switch II), and III (loop region Pro118–Gly126). Regions not involved in significant conformational changes are omitted for clarity. The overall movement of each L subunit toward the L2 dimer interface becomes evident by a 9.4 Å decrease of the distance between the ADP molecules (measured between the N3-atoms of the two adenine bases in the “off state” and the “on state” of L2). Relevant water molecules are shown as red spheres. (B) Identical superposition after a 45° clockwise rotation. (C) Binding of ADP•AlF3 to L2 in the octameric DPOR complex. Residues provided by the second L monomer are indicated by asterisks. The 2Fo–Fc electron density is contoured at 1.5 σ.