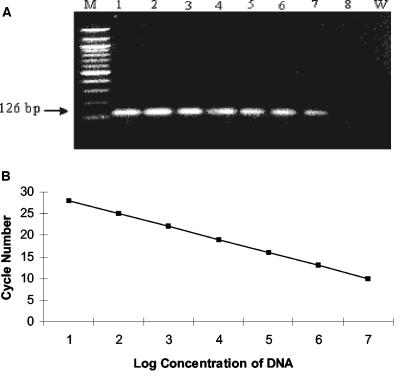
FIG. 1.
Standard curve for K. pneumoniae DNA quantification. (A) Agarose gel electrophoresis of a 126-bp PCR product. Lane W represents the negative control (water). Lane M represents the 100-bp ladder DNA marker; lanes 1 to 8 represent the 10-fold-dilution series of genomic equivalents of 107 copies (lane 1), 106 copies (lane 2), 105 copies (lane 3), 104 copies (lane 4), 103 copies (lane 5), 102 copies (lane 6), 10 copies (lane 7), and 1 copy (lane 8) of purified K. pneumoniae DNA. (B) Plasmid containing one copy of the target sequence was used to construct the standard curve for K. pneumoniae DNA quantification. Serial 10-fold dilutions ranging from 107 copies to 1 copy of plasmid were used for determination of real-time PCR assay detection limits for in vitro sensitivity testing. Cycle number plotted against the log of calculated concentration values resulted in a standard curve with an error of 0.122 and correlation coefficient at unity. Negative-control organisms did not fluoresce above the background signal level.