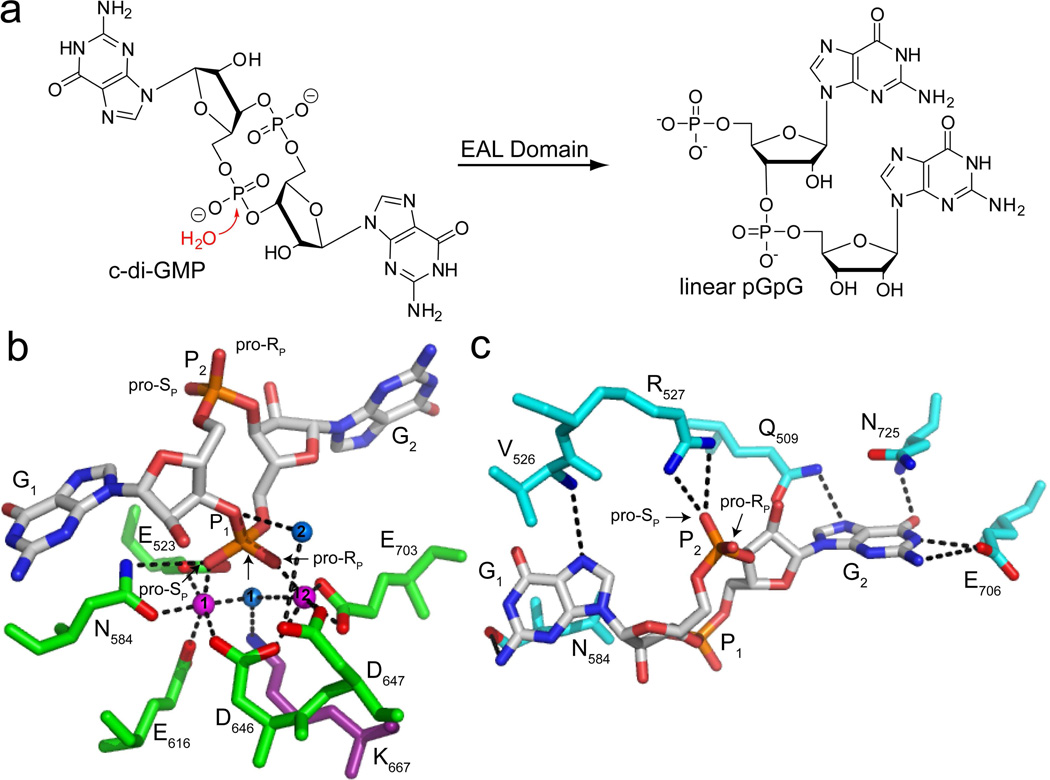
Figure 1.
Recognition and enzymatic hydrolysis of c-di-GMP by EAL domain proteins. (a) Schematic reaction. In the presence of an EAL domain protein, a nucleophilic water molecule attacks one of the c-di-GMP phosphodiester bonds to yield the linear 5′- phosphate product, pGpG. (b) c-di-GMP bound to the active site of the EAL domain protein TBD1265 from Thiobacillus denitrificans (PDB ID 3N3T). c-di-GMP is colored by atom with carbon shown in white, oxygen in red, nitrogen in blue and phosphorous in orange. Water molecules are shown as blue spheres and magnesium ions are shown as magenta spheres. The protein residues in the active site involved in metal coordination are colored in green and oxygens and nitrogens making contacts to c-di-GMP are colored in red and blue, respectively. The highly conserved lysine that stabilizes the negative charge of water 1 and positions it for nucleophilic attack on the scissile phosphate, P1, is shown in purple. Water 2 is predicted to donate a proton to the leaving group. The pro-RP and pro-SP oxygens of P1 and P2 are labeled for sulfur substitution. (c) Recognition of the c-di-GMP bases and the phosphate distal to the site of catalysis, P2 (PDB ID 3N3T). Coloring of c-di-GMP is the same as in (b). Residues in contact with G1 and G2 are colored in cyan and the highly conserved arginine that makes electrostatic contacts to P2 is shown in white. Oxygens and nitrogens of protein side chains are colored as in part (b). Residues involved in G2 recognition are conserved while those involved in G1 recognition are highly variable. The residue corresponding to N725 is most often aromatic (F, W, or Y).