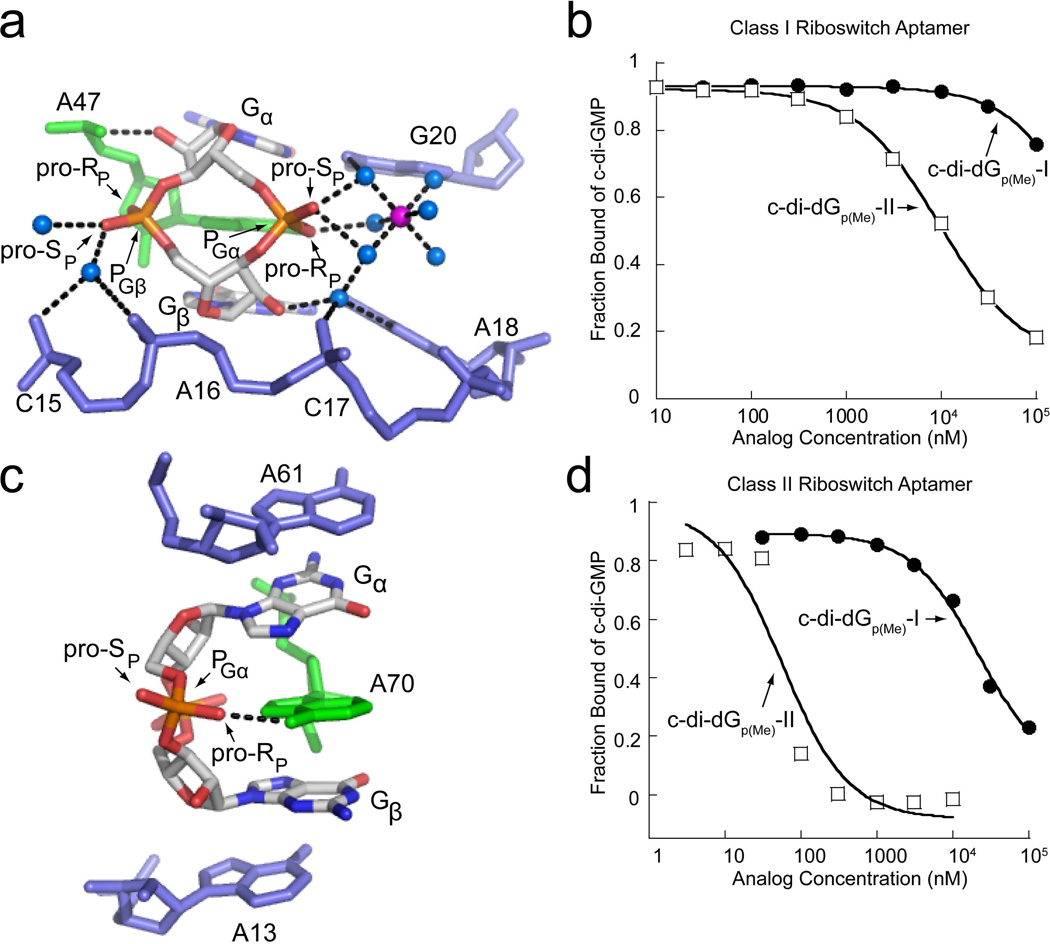
Figure 7.
Backbone recognition of c-di-GMP and binding of methylphosphonate analogs by the class I and class II riboswitch aptamers. (a) c-di-GMP bound to the class I aptamer (PDB ID 3MXH). c-di-GMP is colored by atom with carbon shown in white, nitrogen in blue, oxygen in red, and phosphorous in orange. The highly conserved adenosine that intercalates between the guanine bases of c-di-GMP is colored in green and additional RNA residues are colored in blue. Water molecules are shown as blue spheres and magnesium ions as purple spheres. The pro-RP and pro-SP oxygens are indicated to show the sites of methyl substitutions. Hydrogen bonds are indicated by black dashed lines. (b) Binding curves of methylphosphonate derivatives to the class I riboswitch. The c-di-dGp(Me)-I binding curve is shown as black circles and the c-di-Gp(Me)-II curve is shown as open squares. (c) c-di-GMP bound to the class II aptamer (PDB ID 3Q3Z). Coloring and labeling is the same as in part (a). (d) Binding curves of the methylphosphonate derivatives to the class II riboswitch. Labeling is the same as in part (b).