Fig 5.
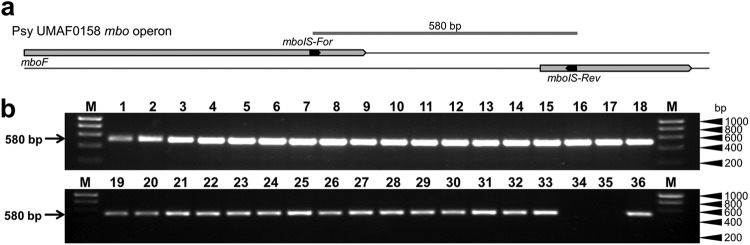
Mapping of the mbo operon insertion point in P. syringae strains. (a) Schematic representation of the PCR amplification strategy for the 3′ end of the mbo operon. Primers mboIS-For/mboIS-Rev (amplicon of 580-bp) were designed between the 3′ end of the mboF gene and the next ORF downstream of the mbo operon. (b) Representative results of amplification using 34 strains that harbor the mbo operon and two non-mangotoxin-producing strains that were used as negative controls. Lanes correspond to the following: 1, P. syringae pv. aptata DSM50252; 2, P. syringae pv. aptata LMG5059; 3, P. syringae pv. aptata LMG5532; 4, P. syringae pv. aptata LMG5646; 5, P. syringae pv. avellanae ISPaVe011; 6, P. syringae pv. avellanae ISPaVe2056; 7, P. syringae pv. pisi HRI203; 8, P. syringae pv. pisi NCPPB1365; 9, P. syringae pv. pisi 1704B; 10, P. syringae pv. syringae 7C6; 11, P. syringae pv. syringae 7B40; 12, P. syringae pv. syringae 7F29; 13, P. syringae pv. syringae 1444-5; 14, P. syringae pv. syringae 1559-9; 15, P. syringae pv. syringae CECT127; 16, P. syringae pv. syringae CECT4429; 17, P. syringae pv. syringae CFBP3388; 18, P. syringae pv. syringae DAR77787; 19, P. syringae pv. syringae EPS17A; 20, P. syringae pv. syringae FF5; 21, P. syringae pv. syringae NCPPB1239; 22, P. syringae pv. syringae Ps-10; 23, P. syringae pv. syringae UMAF0049; 24, P. syringae pv. syringae UMAF0081; 25, P. syringae pv. syringae UMAF0167; 26, P. syringae pv. syringae UMAF0209; 27, P. syringae pv. syringae UMAF0221; 28, P. syringae pv. syringae UMAF1060; 29, P. syringae pv. syringae UMAF2008; 30, P. syringae pv. syringae UMAF2700; 31, P. syringae pv. syringae UMAF2802; 32, P. syringae pv. syringae UMAF4002; 33, P. syringae pv. syringae UMAF6024; 34, P. syringae pv. phaseolicola 1448A; 35, P. syringae pv. syringae B728a; 36, P. syringae pv. syringae UMAF0158; M, molecular size marker (HyperLadder I, Bioline).
