Abstract
In a study aiming to assess bacterial evolution in complex growth media, we evaluated the long-term adaptive response of Escherichia coli MC1000 in Luria-Bertani (LB) medium. Seven parallel populations were founded and followed over 150 days in sequential batch cultures under three different oxygen conditions (defined environments), and 19 evolved forms were isolated. The emergence of forms with enhanced fitness was evident in competition experiments of all evolved forms versus the ancestral strain. The evolved forms were then subjected to phenotypic and genomic analyses relative to the ancestor. Profound changes were found in their phenotypes as well as whole-genome sequences. Interestingly, considerable heterogeneity was found at the intrapopulational level. However, consistently occurring parallel adaptive responses were found across all populations. The evolved forms all contained a mutation in galR, a repressor of the galactose operon. Concomitantly, the new forms revealed enhanced growth on galactose as well as galactose-containing disaccharides. This response was likely driven by the LB medium.
INTRODUCTION
In natural habitats, bacteria are exposed to highly dynamic and complex conditions. In contrast, in artificial environments, such as those found in fermentation systems, this dynamism is often more controlled. However, variations (in space or time) of carbon and energy resources can incite dynamism or heterogeneity, even in such artificial systems.
The complex growth media used in large-scale industrial fermentations may constitute typical examples of such heterogeneous and dynamic environments. Such growth media usually contain inexpensive complex carbon and nitrogen sources that allow good performance (1, 2, 3, 4, 5). Besides the complexity of growth media, other parameters, such as mixing efficiency and perturbation, are also understood to allow variability of local substrate concentrations, pH, temperature, and oxygen levels (6). Such fluctuations may incite dynamism in the perception by the bacteria of their local environment, potentially leading to differential behavior and diversification of the organism that is being grown.
In previous work, locally divergent conditions were suggested to constitute a negative factor for process yields, as bacterial growth and effectiveness were found to be lower in heterogeneous than in homogeneous systems (7). However, in such studies the ability of the bacteria to adapt to environmental conditions (even when they are complex) has been disregarded. Thus, a better understanding of the adaptive response and the mechanistic causes of adaptation and diversification in complex media will contribute to an improvement of our capacities to direct population behavior. This may ultimately improve our abilities to avoid or overcome loss of effectiveness in fermentations.
The fundamental processes involved in the adaptation of bacteria to their environment have been traditionally studied using experimental evolution setups (8), as such experiments allow assessment of the dynamics of adaptation by natural selection within populations. Thus, the growth of a bacterial strain in a defined medium can be maintained for hundreds or thousands of generations. Adaptation can be assessed, since the fitness of evolved forms can be compared, at any time, to that of the ancestral strain (9). Experimental evolution has thus enabled the study of the adaptation of bacteria to the conditions posed by structured versus unstructured habitats (10, 11, 12, 13). The majority of the experiments have used simple limiting/selective conditions (limiting substrates), and only a few studies have included more complex conditions (14), for instance, growth under fluctuations in temperature (15), pH (16), or with combined limited resources (17). As we currently ignore how bacteria may adapt to prolonged growth in complex media, there is a need for studies that address this question (18), which is of major importance to address the effects of the putative coexistence of differentially evolved forms.
Traditionally, fitness of evolved forms has been taken as the average value of a pool of randomly selected evolved forms in competitions separately against their ancestor. However, the variations among these values have often been disregarded. If such variance were considerable, this might indicate the occurrence of heterogeneous responses to the conditions imposed on the evolved populations. This might especially occur in an environment in which the complexity of resources could give space for the occupation of several niches (19). Given the elusive nature of current data on evolution in complex media, this study aims to assess the adaptation of an Escherichia coli strain to growth conditions offered by a complex medium in a long-term sequential batch culture setup. We used Luria-Bertani (LB) medium, in which a multiplicity of energy/carbon sources was present. We thus simulated both spatial and temporal dynamic conditions by including daily propagation into fresh medium and allowing differential oxygen availabilities, including low-oxygen (static oxygen) conditions. The use of LB medium may place limits on the study of physiological characteristics (20); however, we sought to determine if in this complex setup observable fitness diversification would occur at the level of treatment, as well as between or within populations. We thus assessed the evolved forms—versus the ancestor strain—both phenotypically and genetically. The findings point to the occurrence of both common and divergent responses. Substantial evidence was obtained for the occurrence of a major selective genetic event likely having major consequences for the fitness of all evolved forms.
MATERIALS AND METHODS
Bacterial strain and growth medium.
Escherichia coli K-12 strain MC1000 (21) is a facultatively anaerobic organism that was used in this study as the ancestral strain. This strain contains several auxotrophies, being genetically characterized as araD139 Δ(araA-leu)7697 Δ(codB-LacI) galE15 galU strA. Colonies of this strain on LB agar (after 19 h of incubation at 37°C) are morphologically regular, with smooth surfaces and with sizes of approximately 1.5 mm. We used the complex growth medium LB, which consisted of tryptone (10 g/liter), yeast extract (5 g/liter), and NaCl (5 g/liter). Tryptone and yeast extract are both derived from complex sources and have been commonly used as ingredients of fermentation media (2). Tryptone is an enzymatic digest of casein and is rich in tryptophan, large and small peptides, and amino acids. Yeast extract is derived from baker's/brewer's yeast by autolysis at ∼50°C. It has a rich content of amino acids, peptides, water-soluble vitamins, growth factors, trace elements, and diverse carbohydrates. A major carbohydrate of LB medium is the disaccharide trehalose (2, 22).
The long-term evolution experiment.
Seven populations derived from one overnight culture of E. coli K-12 strain MC1000 were propagated under three different oxygen conditions. For this, 0.6-ml aliquots of the washed overnight cultures were introduced into a suite of (i) replicate 100-ml Erlenmeyer flasks containing 60 ml of sterile LB medium (final optical density at 600 nm [OD600], 0.5 ± 0.1) and (ii) 60-ml screw-cap bottles containing 60 ml of sterile LB medium. Two populations (populations 1 and 2) were grown in Erlenmeyer flasks under shaking conditions (200 rpm, oxygen rich; treatment A), three populations (3, 4, and 5) were grown under a cycling environment (oxygen rich, shaking [200 rpm]) in Erlenmeyer flasks, then oxygen-poor static growth in screw-cap bottles, and then returned to shaking (200 rpm) (treatment B); and two populations (6 and 7) were grown in static flasks (oxygen poor; treatment C). All treatment groups were incubated at 37°C during 24 h. Each day, after 24 h (±30 min) of growth in each environment, propagation of the then-stationary-phase cells was performed by applying a 100-fold dilution into fresh medium (final concentration of ∼1 × 107 CFU/ml). In this way, populations went through exponential and stationary phases differentially by treatment, reaching a maximal density of ∼1 × 109 CFU/ml and yielding approximately 7 generations per day. After 150 days, a total of ∼1,000 generations were obtained. Flasks that had been kept under static conditions were mixed before each subsequent transfer, so that the sample would be representative for the diversity in this spatially structured environment. In order to confirm identity and possible visible genomic changes, genomic profiles (based on BOX-A1R-based repetitive extragenic palindromic-PCR [BOX-PCR] and enterobacterial repetitive intergenic consensus-PCR [ERIC-PCR]) were determined every 10 days on several randomly picked clones of each population (23, 24). Throughout all analyses, colony and cell morphologies were also analyzed visually and using microscopy, respectively, supported or not by Gram staining.
Samples taken after every ∼70 generations were kept in glycerol at −80°C as backups in the event of a laboratory mishap. After ∼1,000 generations (over 150 days, excluding mishaps), tubes containing each population (containing the so-called evolved forms) were placed in 20% glycerol at −80°C for further analyses.
Relative/correlated fitness responses and analysis of culture heterogeneity.
The fitness of the evolved forms was measured by competition of each selected form against the ancestor in a crossed experiment. To achieve this, selectable markers were introduced by selecting spontaneous rifampin-resistant (Rpr) mutants of the ancestor as well as each of the evolved forms on 30 μg/ml of rifampin (Rp). Such mutants were then streaked to purity and analyzed by measuring their fitness against the unmutated isogenic form. Only equally fit forms were used in subsequent experiments. Thus, the competitiveness of each of the (marked and unmarked) evolved forms was tested against the (unmarked and marked) ancestor (25, 26). These marker-crossed experiments thus served to assess the level of fitness change versus the ancestor as a presumed result of adaptive evolution.
In detail, prior to the competition experiment, each strain was grown separately in an overnight culture under the condition (environment) for which competition would take place (acclimation phase) (15). After acclimation, the competitors were mixed in 1:1 ratios and placed in the competition environment (competition phase). Each experiment was replicated 6-fold. Over time, samples were taken and plated in duplicate onto LB agar with or without Rp supplementation to estimate the population size of each competitor. After overnight incubation of the plates, the mean numbers of colonies on the Rp-containing plates were subtracted from those on plates without rifampin. Both data sets were then used to obtain estimates of the numbers of both competitors (27).
Malthusian parameters (m) for each competitor were calculated as follows: m = ln[(density at end of competition)/(density at time zero of competition)] (12). Relative fitness values were obtained for the evolved forms in competition with the ancestral strain under the specific environment. The relative fitness, W, of an evolved form (E) relative to the ancestor (A) was calculated as the ratio of both Malthusian parameters (12): W = mE/mA. The evolved forms from treatment B were competed, including daily transfers from shaking to static conditions, for 3 days. This way, competitors were exposed to both transitions of this treatment. Acclimation took place under aerobic conditions (25).
Correlated fitness analysis (growth in alternative environments) was performed in order to examine the specificity of adaptation with respect to each treatment (presence of specialists or generalists). Thus, following the method described above, clones from each experimental line were subjected to the other two experimental treatments in competition with the ancestor (10, 16, 17).
Phenotypic diversity.
The metabolic potentials of each selected evolved form as well as the ancestor were analyzed in duplicate on the 95 substrates of GN2 plates (Biolog Inc., Hayward, CA). The absorbance values at 600 nm were measured using a microplate reader (VersaMax microplate reader; Molecular Devices Corp.). For each strain, cells were pulled into phosphate-buffered saline (PBS; pH 6.5), and the suspension was set at an OD600 of 0.04. Then, 100 μl was transferred to each well of the GN2 plates, including one blank. The plates were incubated at 37°C and read after 24, 48, and 72 h. Average absorbance values were used for cluster analysis after scoring, as follows: 0, no catabolism; 1, catabolism (weak responses included). The cutoff point between 0 and 1 was an OD of 0.2 (28).
Specific results were independently confirmed in test tubes with M9 minimal medium (1× M9 salts, 2 mM MgSO4, 0.1 mM CaCl2, 0.01% yeast extract) supplemented or not with 0.4% of specific carbon sources. Bacterial growth was followed during 24 h, and OD600 values were recorded. Moreover, time zero and late-exponential-phase samples were plated on LB agar for CFU counting.
We included cell length as a phenotypic property to observe diversification. Cells of each treatment, next to the ancestor, were taken directly from the endpoint populations (60 cells per population) and observed under a microscope (Axiostar Plus HBO 50Ac; Zeiss). Cell length was further calculated by using AxionVision software v.4.8 2.0 (Carl Zeiss MicroImaging GmBH).
Comparative genome sequencing and screening for genetic diversity.
The endpoint populations of each evolved strain, next to the ancestor, were streaked onto LB agar plates. The selected colonies were used to found cultures for DNA isolation using the Gnome DNA isolation kit (MP Biomedical). Following extraction, the genomic DNA was fragmented using a Bioruptor (Diagenode) for 10 cycles of 30 s on/30 s off. Then, they were converted into paired-end Illumina sequencing libraries by using the NEBNext Quick DNA sample prep master mix set 2 (NEB, Ipswich, MA), according to the manufacturer's protocol. Multiplex adaptors (MA; multiplexing sample preparation oligonucleotide kit; Illumina, San Diego, CA) were used in the adaptor ligation. Library enrichment was carried out in two amplification steps using primers and indices described earlier (29). Genome libraries with individual indices were pooled, to contain 10 or 11 genomes per pool, and sequenced, with one lane per pool. Sequencing was performed by 107 cycles on a HiSeq2000 instrument (Illumina), yielding an average of >20 million reads of, on average, 91 bases each (subtracting the index).
To identify all mutations in the evolved strains versus the ancestor, we used Breseq v.014 (30). This is a computational pipeline for the analysis of short-read sequencing data. Thus, we used the sequenced and annotated GenBank file of the genome of E. coli K-12 MG1655 (GenBank U00096.2 [31]) as a reference and mapped all high-quality reads of the sequenced genomes. The quality of the reads was determined using Biopieces (http://www.biopieces.org) and BWA (32). Mutations were confirmed by identification of single-nucleotide polymorphisms (SNPs) and deletion-insertion polymorphisms (DIPs) by using SAMtools (33). Differences found in our ancestral strain from the reference E. coli K-12 MG1655 genome were considered the “genetic property” of our ancestor. In this manner, unique differences between the evolved and ancestor strains could be visualized. After obtaining the positions and descriptions of all mutations, the putative functions of the gene hits were assessed using DAVID v.6.7 (functional annotation tool) at default settings (34, 35). The resulting function terms were further grouped by similarities to determine the enrichment (geometric mean of all modified Fisher's exact test P values) of biological processes. High enrichment scores were taken into account, but genes with a low enrichment score (<1.3) were also analyzed, as recommended by the developer.
RESULTS AND DISCUSSION
Adaptation of evolved forms.
After 150 days of daily propagation under three different oxygen conditions, we obtained cultures of evolved E. coli MC1000 that had undergone approximately 1,000 generations. The three different environments used involved cultures growing in flasks with LB medium under constant shaking conditions (treatment A, oxygen rich), cycling between shaking and static growth (treatment B, cyclic oxygen rich and oxygen poor), and static growth (treatment C, oxygen poor).
The final populations at day 150 were diluted and plated on LB agar in order to produce single colonies representing evolved forms. Colony polymorphism was clearly observed across all evolved populations. At this point, forms were classified as (i) small colonies (“a” type), with diameters of <1 mm, (ii) large/rough/irregular colonies (“b” type), with diameters of >1 mm, and (iii) large/smooth/regular colonies (“c” type), with diameters of >1 mm. Treatments A and B showed colony types a, b, and c in all populations. In contrast, treatment C showed only types b and c colonies. A total of 19 colonies were selected across all populations. These were subjected to all subsequent analyses of phenotype and genotype. Throughout the experiment, genomic fingerprinting analysis did confirm the identity of all colonies as E. coli MC1000, as evidenced from their characteristic patterns, i.e., similar to the ancestor (data not shown).
To assess the overall fitness of each of the evolved forms, we evaluated their fitness by competing them against the ancestor. These marker-crossed experiments revealed consistencies in the fitness results between the use of marked and unmarked evolved forms, as there were no significant differences between any set of results (P > 0.05). Also, marked and unmarked ancestors did not show any significantly different fitness response when competed against each other. Thus, the marker-crossed experiments allowed us to assess any fitness changes in each of the evolved forms versus the ancestor.
The ancestral E. coli MC1000 strain grew and survived well in both the oxygen-rich and oxygen-limited LB cultures. However, it was outcompeted by all endpoint evolved forms in all environments (significant at P < 0.05) (Table 1). The forms that had evolved under treatments A and C revealed large fitness increases in these environments compared to the ancestor, whereas the forms from treatment B showed smaller increases. The raised fitness values observed in the more stable environments suggested that such noncycling conditions may have given better niche space or time for fitness-enhancing changes to be fixed.
Table 1.
Direct and correlated fitness responses after 150 days of culturea
| Response type and treatment group | Conditionb | Wc (mean ± SEM) |
P value for fitness comparison between: |
||
|---|---|---|---|---|---|
| Evolved vs ancestord | Direct vs correlatede | Different populationsf | |||
| Direct fitness responses | |||||
| A | 1.162 ± 0.03690 | 0.001 | 0.002 | ||
| B | 1.114 ± 0.00991 | <0.001 | 0.058 | ||
| C | 1.351 ± 0.03153 | <0.001 | 0.029 | ||
| Correlated fitness responses | |||||
| A | Cl | 1.059 ± 0.02983 | <0.05 | ||
| A | St | 1.073 ± 0.01337 | <0.05 | ||
| B | Sh | 1.129 ± 0.02232 | 0.696 | ||
| B | St | 1.279 ± 0.02432 | <0.001 | ||
| C | Sh | 1.103 ± 0.00919 | <0.001 | ||
| C | Cl | 0.978 ± 0.01332 | <0.001 | ||
Fitness results were obtained after competition between the selected isolates and the ancestor. Significant differences (P < 0.05) are shown in boldface.
Correlated fitness competitions occurred under the following conditions: Sh, shaking; Cl, cycling; St, static.
W represents the mean of all the competitions for each treatment.
Based on two-tailed t test. Ancestor with a null hypothetical value = 1.0.
Comparison between correlated and direct fitness responses.
Based on one-tailed t test performed on replica populations belonging to the same treatment group (for treatment C, a one-way ANOVA was employed).
Competition of evolved forms with the ancestor in the alternative environments (correlated fitness) was considered to reveal the specificity of adaptation for either the medium or oxygen conditions. The results showed that populations from both treatments A and C grown in the two alternative environments revealed relatively lower fitness increases than those in their own environment (P < 0.05) (Table 1). This suggested the occurrence of possible trade-offs in these forms, decreasing their relative competitiveness enhancement in the alternative environments and increasing it under their “own” conditions. On the other hand, the treatment B forms outcompeted the ancestor under static conditions in a stronger manner than in their own environment, showing no significant difference when grown under shaking conditions (P < 0.05) (Table 1). This suggested the presence of more generalist forms.
To observe the fitness potential of the total endpoint populations versus the ancestor (and a possible existence of interactions), samples of each endpoint population (all clones together) were competed 1:1 against the ancestor under their “own” conditions. Results showed the fitness increase values to be similar to the averaged values of the individually competed forms for treatments A and C (P > 0.05) (data not shown). A striking difference was observed in the populations of treatment B, which revealed average fitness increases of 18% compared to the average fitness of the individual clones. This increase suggested that genotype-genotype interactions (e.g., yielding cross-feeding) may have played a role (19). Adaptive changes in the forms evolving under treatment B might have resulted in the emergence of generalists that are able to cope with the oxygen-rich or oxygen-limited conditions and/or the transitions between these two conditions. When living together, the presence of forms that are fit under one of the conditions might diminish the impact of the change to conditions of another form (which may thus be coselected) (36). This contention was consistent with the positive response of forms from treatment B to the correlated fitness analysis.
Heterogeneity of fitness values within and between populations.
A closer observation of the relative fitness values of the evolved forms highlighted a remarkable fitness diversity between replicate populations as well as within populations. The former comparison (Table 1) indicated that the average fitness gains versus the ancestor were different between the replicate populations of treatments A and C (P < 0.05; standard error of the mean [SEM] > 0.03), whereas they were similar for those of treatment B (P = 0.06). This indicated that, in each replicate population of treatments A and C, forms may have coemerged that evolved along a different path, modulating their cellular machineries in response to the environment in different ways. For treatment B, a more coherent evolutionary path may have been followed. We then assessed the within-population variations in the fitness gains in treatment groups A through C. Nested analysis of variance (ANOVA) revealed a wide dispersion of values in all populations of treatment groups A and C (PASW-SPSS, 18.0.3; P < 0.05), with variance values of 0.0081 and 0.0039, respectively. In contrast, clones from treatment B showed more similar relative fitness values between each other (variance, 0.0008).
Phenotypic characteristics confirmed diversification and also paralleled adaptive responses.
The cell sizes of the evolved forms versus the ancestor were determined using cell length (in μm) as a proxy. In comparison to the ancestral strain, the evolved forms from treatment groups A and B showed increased cell lengths, whereas those of treatment C revealed decreased cell lengths (P < 0.01). Specifically, the ancestor cells had an average length of 1.48 μm (±0.182), whereas those of treatments A and B had lengths of 1.82 μm (±0.251) and 1.66 μm (±0.349), respectively. In contrast, cells of treatment group C showed about 18% decreased cell lengths relative to the ancestor (1.22 ± 0.189 μm).
We surmised that the increased cell sizes observed in treatment groups A and B might be attributable to the oxygen-rich conditions used. Possibly, the harvested evolved forms had become phenotypically well adapted to the utilization of the complex substrates of LB under oxic conditions, resulting in larger cells (37). Considering treatment C, reduction of cell lengths of the evolved forms might be related to processes under these conditions, in which σS-controlled gene products may generate changes in the cell envelope and overall morphology (38).
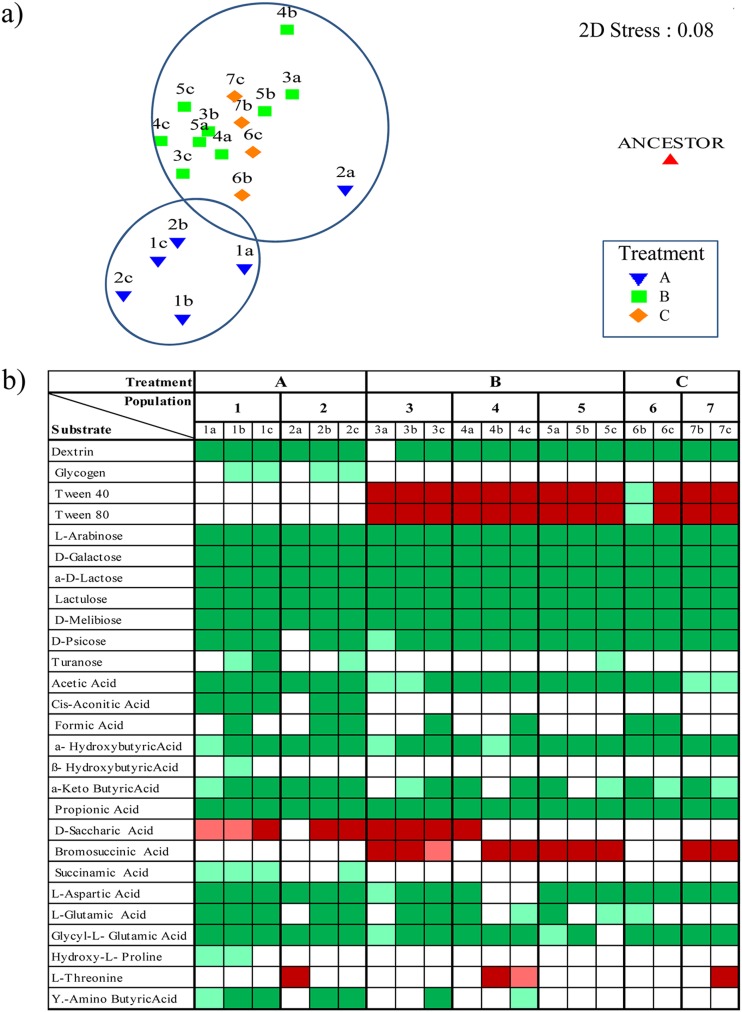
For all evolved forms, the Biolog phenotypic arrays revealed weak responses to most substrates after 24 h. After 48 h, avid consumption of most of the utilizable substrates became evident, whereas after 72 h the data were confirmed. Replicate assessments were consistent, with, on average, less than a 6.3% difference between these. Expectedly, the ancestor strain metabolized 44.2% of the substrates, which was consistent with previous work with E. coli K-12 (39). The evolved forms of treatment group A metabolized, on average, 63.5%, and those of treatment groups B and C metabolized 54.7% and 56.3% of the substrates, respectively. Multidimensional scaling (MDS) was performed to observe similarities among the responses of the evolved forms and compare them to the responses of the ancestor (Fig. 1a). All evolved forms clustered consistently per treatment (except for 2a, i.e., from population 2 of treatment A) and away from the ancestor (stress, 0.08). Interestingly, the responses of forms from treatments B and C resembled each other. These two treatments shared the oxygen limitation condition, suggesting that comparable metabolic pathways could have been affected in the evolved forms.
Fig 1.
Phenotypic microarrays results (with Biolog GN2 plates). (a) Nonmetric multidimensional scaling (MDS) plot displaying similarity between all selected clones from the three treatment groups in response to growth in the 95 substrates of the GN2 Biolog plates. The position of the ancestor is shown. The plot was derived using a Bray Curtis similarity matrix, and the stress level is indicated. Two main clusters were observed (circles). (b) Differential Biolog profile (relative to the ancestor) of the selected clones from treatment groups A, B, and C. Green boxes are novel substrates metabolized after 150 days of growth under each regimen, red boxes indicate lost ability to metabolize the substrate. Boxes that are light green or light red indicate substrates with a weak response (i.e., they either changed to be consumed [green] or did not [red] in comparison with the ancestor). Blank boxes indicate no change occurred.
Then, a matrix with a total of 24 informative substrates (those that had shown differential responses of evolved forms relative to the ancestor) was then constructed (Fig. 1b). Using this matrix, both differential responses (even within populations) and consistent ones (even across all treatments) were noted. The finding of common responses in evolved forms across all treatments (mostly metabolic upshifts) was striking. A closer examination using the level of KEGG networks revealed that there was consistency in the metabolism of carbohydrates. In particular, several early (at 24 h) upshifts were related to the galactose metabolic pathway. With shunting into this pathway, disaccharides with a galactose moiety, such as α-d-lactose, d-meliobiose, and lactulose, can be quickly consumed. This metabolic upshift was exactly what we found. Moreover, a consistent upshift in l-arabinose utilization (which provides an entrance to the pentose phosphate pathway) was also observed.
Thus, a consistent adaptive response might have been triggered by a factor common to all treatments, most likely (and particularly the sugar components of) the LB medium used. Considering this fact, we decided to assess the growth of the evolved forms and the ancestor in M9 minimal medium by using the disaccharides found as potential selective factors (Biolog phenotypic array data) as sole carbon sources (d-lactose, d-melibiose, and d-galactose). We also used l-arabinose and d-trehalose to confirm positive metabolism of the evolved forms. All results were positive; in other words, all populations inoculated at 103 CFU/ml reached concentrations of 107 CFU/ml in late exponential phase (after 9 h), whereas the ancestor was not able to grow (except in d-trehalose, where it reached 106 CFU/ml after 9 h). LB medium is known to contain little glucose, which disappears after 2 h of growth of E. coli (1). However, it offers several sugars (next to alcohols and organic acids), i.e., trehalose, arabinose, lactose, and maltose, in significant levels (22). In fact, the level of the major sugar component of LB medium (trehalose) has been reported to be as high as 17% (1, 22, 40). Trehalose (with an α,α-1,1-glucoside bond between two α-glucose units) may thus offer the major entrance to the glycolytic pathway for organisms growing in LB medium. Moreover, alternative pathways, such as the galactose and pentose phosphate pathways, may serve as sources of intermediates (41, 42). Our findings suggested an association of the consistent metabolic upshifts in the evolved forms with the selection of metabolic strategies converging to glycolysis.
Additionally, these metabolic tests revealed that forms even from within the same population may have different metabolic profiles. For example, when population 2 (from treatment A) was grown in d-galactose, a remarkable difference in the response of form 2a compared to the other forms could be easily observed. Compared to forms 2b and 2c, form 2a showed delayed metabolism of d-galactose, reaching only 1.0 × 105CFU/ml after 9 h (see Fig. S1 in the supplemental material). From the differential matrix (Fig. 1b), we also observed that the “a” form types from populations 1, 2, and 3 (1a, 2a, and 3a) were metabolically unique compared to the other two types in the same populations. Some heterogeneous responses were seen for substrates belonging to related pathways, e.g., the alanine, aspartate, and glutamate metabolic pathway (l-aspartic acid, l-glutamic acid, and glycyl l-glutamic acid) and pathways for several related carboxylic acids.
Genomic changes and natures of affected genes.
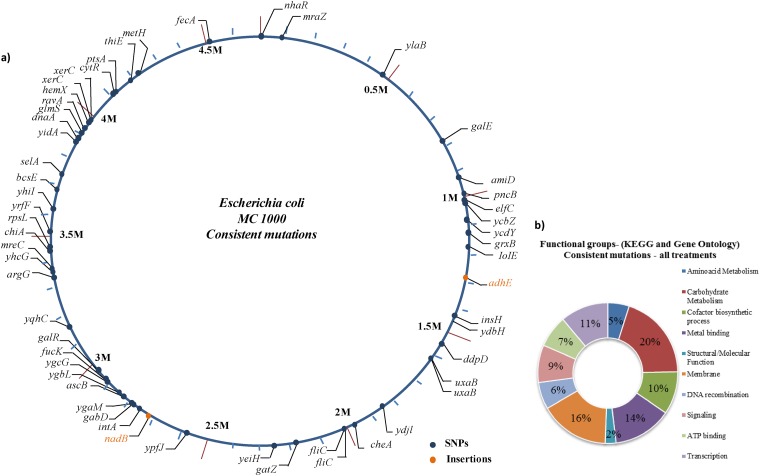
Analyses of the BOX-PCR and ERIC-PCR profiles of all evolved forms did not show any differences from those of the ancestor. To detect genomic differences, we then performed Illumina-based sequencing of genomic DNA from each of the selected evolved forms (totaling 20 genomes, including the ancestor). Overall, the SNP analysis methods SAMtools and Breseq revealed the same mutations per evolved form. Assuming all synonymous mutations to be neutral and all nonsynonymous mutations to be deleterious, neutral, or advantageous, we calculated the ratio of the number of nonsynonymous substitutions (dN) to that of synonymous substitutions (dS) (using only coding regions). A dN/dS ratio of >1 implies that the changes incurred are positive and related to adaptation (43). The dN/dS ratios for treatments A and C were 2.5 and for treatment C it was 2, thus indicating that the changes found were, overall, beneficial for adaptation. Compared to other experimental evolution studies, in which whole-genome sequencing has been applied, the numbers of nonsynonymous mutations were rather high (18). However, the former data were based on studies in defined media, and this is the first report in which differences in medium complexity (combined with variations in oxygen availability) were included. Interestingly, a total of 55 nonsynonymous mutations occurred in parallel in all evolved forms in all treatment groups (Fig. 2a). In addition, 23 mutations consistently occurred in intergenic (presumably noncoding) regions (see Table S1 in the supplemental material). Parallel mutations found in independent evolving lines have been reported in long-term experiments (30, 44, 45, 46, 47). These types of parallel changes have been characterized as beneficial targets of selection.
Fig 2.
Consistent mutations (single-nucleotide polymorphisms [SNPs] and deletion/insertion polymorphisms [DIPs]) found after the long-term experiment. (a) A total of 55 intragenic nonsynonymous mutations in all treatment groups were consistent and were distributed all around the genome. (b) Functional groups of all consistent mutations found with the DAVID v.6.7 functional annotation tool.
Bioinformatics-based results (using DAVID v.6.7) suggested that the major functional genes (functional groups) that had been consistently affected were genes involved in carbohydrate metabolism (20% of the total). Another 16% of affected genes were related to membrane processes, and 14% were related to processes involving metal binding (based on KEGG and Gene Ontology analysis [DAVID v.6.7]) (Fig. 2b). Some other important functional groups were also consistently affected, among which transcriptional regulator genes (i.e., galR, cytR, dnaA, and ravA). Consistency in changes of genes related to carbohydrate metabolism correlated well with the improved (and also consistent) responses to carbohydrates of the Biolog plates. Our ancestral strain was unable to utilize galactose, presumably because its genotype has a mutation in one of the gal operon genes (galE15) that results in a change of an amino acid (Ser to Phe). GalE catalyzes a hydride transfer in the interconversion of UDP-galactose and UDP-glucose as part of galactose catabolism. Interestingly, we found a consistent point mutation in our evolved forms that reverted this mutation to the original state of E. coli K-12, able to consume galactose as sole carbon source (48). Moreover, a galR point mutation located in nucleotide 2975171 of the genome (C→T), changing the codon TCT to TTT, was consistently observed. As a result, a polar amino acid (serine) was replaced with a nonpolar one (phenylalanine). GalR is the major repressor of the gal operon (galETKM). ΔgalR mutants have been found to incite induction of the gal operon (3-fold higher expression than the wild type) in the presence of galactose (42, 49). The consistent galR mutation and the consistent metabolic upshift observed in the Biolog results provide ammunition for the hypothesis that these are the basis of the consistently enhanced fitness in all evolved forms. Apparently, E. coli MC1000 cells evolving in LB medium that have undergone a switch to derepression of the galactose operon are amenable to selection by the force exerted by disaccharides that are present in the medium.
Similarly, mutations were found in genes involved in metal binding. LB is a medium that contains very small amounts of divalent cations (Mg2+ and Ca2+) (50). Given the importance of acquisition of sufficient metals by organisms evolving in LB medium, this would clearly agree with the presumed importance of fitness-enhancing changes in genes involved in metal binding. Moreover, the changes found in membrane-related genes might also relate to this hypothesis, as well as to the idea that cells may have evolved to facilitate the uptake of carbon/energy sources through their membranes.
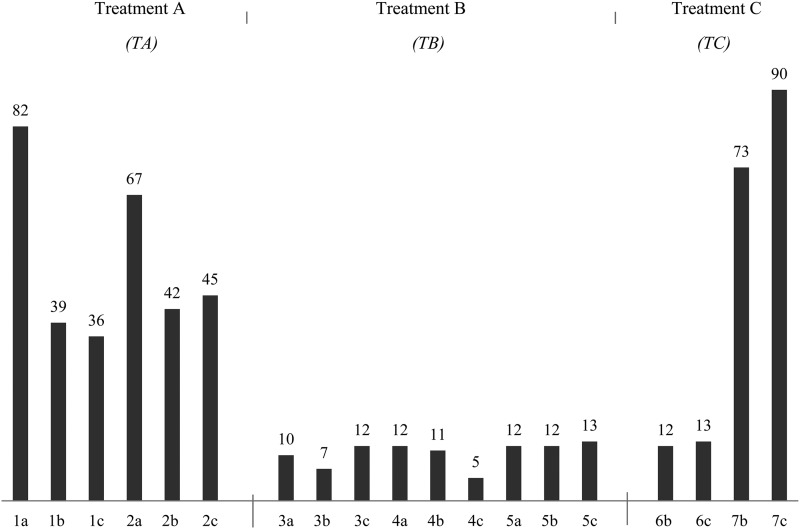
Moreover, a suite of nonconsistent (not occurring in all forms) mutations was found in a nonrandom fashion (Fig. 3). The highest numbers of mutations (intragenic and intergenic) were found in treatments A and C, with an average 52 and 47 mutations (P > 0.05), respectively. In contrast, treatment B showed a lower average number (10) of mutations (treatment A versus B, P < 0.01; treatment A versus C, P > 0.05; treatment B versus C, P > 0.05, two-tailed t test). The numbers of mutations were found to correlate well with the levels of fitness increase. First, evolved forms from treatment B showed the lowest fitness improvements and the lowest numbers of genetic changes. Second, in evolved forms of treatment groups A and C, the numbers of nonconsistent mutations were high, whereas the raised dN/dS ratios indicated that the numbers of positively selected mutations were higher, which in an overall fashion would lead to their higher fitness.
Fig 3.
Mutations (SNPs and DIPs) nonconsistent among all populations. The total number of nonsynonymous single-nucleotide substitutions, insertions, and deletions (nonconsistent among all treatment groups) found after sequencing using Illumina Hiseq2000 are shown. Intragenic and intergenic mutations are included. There was a significant difference in the number of mutations between treatments A and B (TA versus TB, P < 0.01). There was no significant difference when treatment C was compared to the other two treatments (TA versus TC, P > 0.05; TB versus TC, P > 0.05; two-tailed t tests).
To observe genomic diversification at the population level, the numbers of mutations were compared between replicate populations. No significant differences were found in the average numbers of mutations between replicate populations of treatment groups A and B. However, the difference between the two populations of treatment C, i.e., 69 mutations, was remarkable. The environmental heterogeneity, providing a spatial structure for treatment C, may have led to higher diversification (51). Another finding was that unique mutations (not found in other forms of the same treatment group or other treatment groups [see Table S2 in the supplemental material) were observed. Forms 1a, 2a, and 7c revealed high numbers of unique mutations, i.e., 55, 56, and 21, respectively.
Functional annotation analysis of nonconsistent mutations.
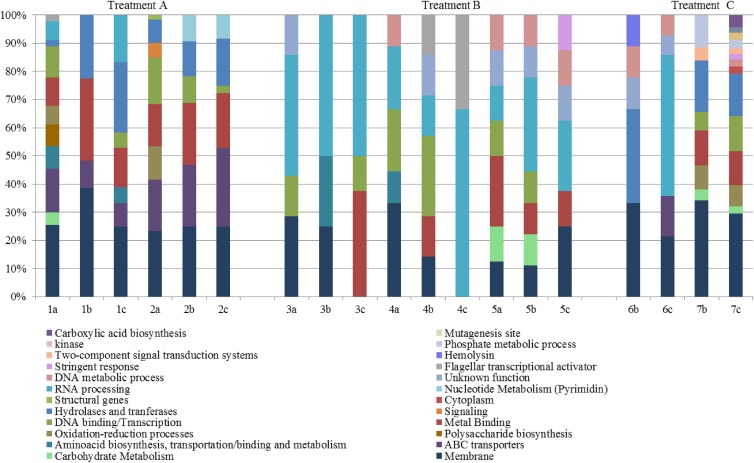
Results of the functional annotation analysis for all nonconsistent mutations (Fig. 4) showed similarities between treatments as well as treatment group particularities. First, the evolved forms of treatments A and C shared several common mutations. Strikingly, the functional group with the largest number of genes affected with mutations was the group “membrane processes.”
Fig 4.
Functional groupings of the nonconsistent mutations among all population mutations. The graph shows the different functional categories (using KEGG and GO terms) of the genes affected that displayed no consistent mutations (to any treatments). The percentages correspond to the amount of mutated genes that fell into each functional category. Enrichment analysis was performed with DAVID v.6.7.
A second functional group with mutations common to these two treatments was that concerned with hydrolase and transferase proteins. Treatment A apparently incited consistent mutations in the ABC transporter genes thkH, ompA, mppA, ssuB, ssuC, ompF, macB, and rbsA across all clones. Some of these transporters are modulators of the passage of solutes like sugars, ions, and amino acids. Also, treatment A consistently showed mutations in regulatory genes involved in central metabolism (arcA and arcB). Mutations in these two-component regulatory system genes (under aerobic conditions) have been found to be responsible for upregulation of genes whose products are involved in the tricarboxylic acid cycle (52). Treatment group B had the lowest number of mutated genes involved in transcription regulatory processes, but it showed consistency with respect to the gene rpoA. This gene encodes the α-subunit of RNA polymerase and was found to carry a single-base transition, leading to a substitution of Asn for His at position rpoA294. In general, an overall comparison of functional annotations between populations of the same treatment group indicated similarities in the functions affected by mutations. Only treatment group C showed less commonality between its two populations.
The unique mutations found in forms 1a, 2a, and 7c involved genes related to processes such as oxidation-reduction (trxC, aidB, and narZ), carbohydrate metabolism (aldA, leuA, fucO, and tktA), polysaccharide biosynthesis (wzzE, rfc, ispA, rfaG, mppA, and plsC), flagellar transcriptional activation, and DNA binding processes. Phenotypically, these forms appear to have different metabolic profiles compared to their coevolved partners, and these mutations and their epistatic effects may cause these particularities.
An analysis of the extent to which individual genetic variation contributes to changes in phenotype is difficult, due to possible epistatic interactions (53). Conrad et al. in 2009 suggested that an analysis of important regulatory regions needs to be performed, and the impact of changes in such regions has to be evaluated by mutations of wild types (54). Further work will analyze such effects in our evolved forms.
Conclusion: the adaptive response to LB medium is related to selection for galactose derepression.
In this study, we evaluated the adaptation of E. coli MC1000 following evolution for ∼1,000 generations along parallel evolutionary paths in a complex environment defined by medium (LB) and oxygen condition. Moreover, we described, in an integrative way, the effect of environment, i.e., the presence or absence of oxygen and shifts therein of the growth medium, on adaptation during prolonged growth of the organism. Substantial mutational and metabolic differences could be distinguished in the endpoint evolved forms, even within populations. The number of potential niches offered by the medium was likely large enough to allow such diversification. This is in contrast with previous studies performed in simpler (defined) growth media, as these studies found a lack of diversification (55, 56). Populations that have evolved under temporal environmental variations have less space to diversify than those that have evolved in structured environments. However, possible early fixation of changes might have occurred, leading to the establishment of generalists with possible interactive strategies and an ability to handle both oxygen conditions.
Collectively, the above phenotypic and genomic data indicate that all evolved forms modulated their metabolism toward the utilization of particular carbohydrates. A major consistent metabolic upshift occurred in the galactose utilization pathway, resulting in the enhanced utilization of particular disaccharides that feed into this pathway. It is plausible that this enhanced metabolism ultimately results in enhanced metabolic flux (shunting into glycolysis and the Krebs cycle); in other words the amount of substrate used per unit of time is increased in all evolved forms.
Previous studies of genomic changes in independent E. coli populations grown under different conditions already identified several consistent adaptive mutations in regulatory genes (57, 58). The positive reversion of the galE mutation and the mutation in the galR repressor might be involved in the fitness increase of our evolved forms as a key strategy to obtain carbon and energy. Clearly, unique mutations can be responsible for the variability among the populations, possibly being proxies for genetic drift.
The parallel adaptive responses suggested that, even though environment drove adaptation, the medium played the key role in the adaptive response. Thus, E. coli modulates its metabolism to cope with the scarcity of easily available carbon and energy sources like glucose in LB medium, and it can respond by activating alternative pathways converging to glycolysis. On the basis of our data, we have inferred that key adaptive responses to diverse nutritional sources, in this case, different types of small sugars, are major determinants of evolutionary adaptions, whereas further diversification may occur as a consequence of medium complexity.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Department of Biology, Section of Microbiology, University of Copenhagen, for its support and contributions to this paper. We also acknowledge the valuable help of Lars Hansen for carrying out the Illumina sequencing runs. We thank all colleagues from the Microbial Ecology Department of the University of Groningen for their input and support.
Financial support was provided by the ERANET IB program of NWO (2nd).
Footnotes
Published ahead of print 30 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02920-12.
REFERENCES
- 1. Baev MV, Baev D, Janeso A, Campbell JW. 2006. Growth of Escherichia coli MG1655 on LB medium: monitoring utilization of sugars, alcohols and organic acids with transcriptional microarrays. Appl. Microbiol. Biotechnol. 71:310–316 [DOI] [PubMed] [Google Scholar]
- 2.Pasupuleti VK, Demain A. Protein hydrolysates in biotechnology. Springer; Dordrecht, Heidelberg, Germany: 2010. [Google Scholar]
- 3. Silva L, Passarinha F, Sousa F, Queiroz JA, Domingues FC. 2009. Influence of growth conditions on plasmid DNA production. J. Microbiol. Biotechnol. 19:1408–1414 [DOI] [PubMed] [Google Scholar]
- 4. Tripathi N, Shrivastva A, Biswal KC, Rao PVL. 2009. Optimization of culture medium for production of recombinant dengue protein in Escherichia coli. Ind. Biotechnol. 5:179–183 [Google Scholar]
- 5. Wang D, Li Q, Yang M, Zhang Y, Su Z, Xing J. 2011. Efficient production of succinic acid from corn stalk hydrolysates by a recombinant Escherichia coli with ptsG mutation. Process Biochem. 46:365–371 [Google Scholar]
- 6. Enfors SO, Jahic M, Rozkov A, Xu B, Hecker M, Jürgen Krüger BE, Schweder T, Hamer G, O'Beirne D, Noisommit-Rizzi N, Reuss M, Boone L, Hewitt C, McFarlane C, Nienow A, Kovacs T, Trägårdh C, Fuchs L, Revstedt J, Friberg PC, Hjertager B, Blomsten G, Skogman H, Hjort S, Hoeks F, Lin HY, Neubauer P, van der Lans R, Luyben K, Vrabel P, Manelius A. 2001. Physiological responses to mixing in large scale bioreactors. J. Biotechnol. 85:175–185 [DOI] [PubMed] [Google Scholar]
- 7. Lara AR, Galindo E, Ramírez OT, Palomares LA. 2006. Living with heterogeneities in bioreactor, understanding the effects of environmental gradients on cells. Mol. Biotechnol. 34:355–381 [DOI] [PubMed] [Google Scholar]
- 8. Nakatsu C, Korona R, Lenski R, de Bruijn F, Marsh T, Forney L. 1998. Parallel and divergent genotypic evolution in experimental populations of Ralstonia sp. J. Bacteriol. 180:4325–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bennett A, Hughes BS. 2009. Microbial experimental evolution. Am. J. Physiol. Regul. Integr. Comp. Physiol. 297:17–25 [DOI] [PubMed] [Google Scholar]
- 10. Bennett A, Lenski RE, Mittler J. 1992. Evolutionary adaptation to temperature. I. Fitness responses of Escherichia coli to changes in its thermal environment. Evolution 46:16–30 [DOI] [PubMed] [Google Scholar]
- 11. Finkel SE, Kolter R. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. U. S. A. 96:4023–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lenski RE, Rose MR, Simpsom SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315–1341 [Google Scholar]
- 13. Rainey PB, Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 32:69–72 [DOI] [PubMed] [Google Scholar]
- 14. Barrett RDH, MacLean RC, Bell G. 2005. Experimental evolution of Pseudomonas fluorescens in simple and complex environments. Am. Nat. 166:470–480 [DOI] [PubMed] [Google Scholar]
- 15. Leroi A, Lenski RE, Bennett A. 1994. Evolutionary adaptation to temperature. III. Adaptation of Escherichia coli to a temporally varying. Evolution 48:1222–1229 [DOI] [PubMed] [Google Scholar]
- 16. Hughes BS, Cullum AJ, Bennett A. 2007. Evolutionary adaptation to environmental pH in experimental lineages of Escherichia coli. Evolution 61:1725–1734 [DOI] [PubMed] [Google Scholar]
- 17. Cooper TF, Lenski RE. 2010. Experimental evolution with E. coli in diverse resource environments. I. Fluctuating environments promote divergence of replicate populations. BMC Evol. Biol. 10:11 doi:10.1186/1471-2148-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dettman JR, Rodrigue N, Melnyk H, Wong A, Bailey SF, Kassen R. 2012. Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Mol. Ecol. 21:2058–2077 [DOI] [PubMed] [Google Scholar]
- 19. Kassen R, Rainey PB. 2004b. The ecology and genetics of microbial diversity. Annu. Rev. Microbiol. 58:207–231 [DOI] [PubMed] [Google Scholar]
- 20. Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Casadaban MJ, Cohen SN. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207 [DOI] [PubMed] [Google Scholar]
- 22. Hanko VP, Rohrer JS. 2000. Determination of carbohydrates, sugar alcohols, and glycols in cell cultures and fermentation broths using high-performance anion-exchange chromatography with pulsed amperometric detection. Anal. Biochem. 283:192–199 [DOI] [PubMed] [Google Scholar]
- 23. Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Versalovic J, Schneider M, de Brulin FJ, Lupski JR. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25–40 [Google Scholar]
- 25. Bennett E. 1996. Evolutionary adaptation to temperature. V. Adaptation of Escherichia coli to a temporally varying environment. Evolution 50:493–503 [Google Scholar]
- 26. Gagneux S, Long CD, Small P, Van T, Schoolnik GK, Bohannan BJM. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944–1946 [DOI] [PubMed] [Google Scholar]
- 27. Enne VI, Delsol A, Roe JM, Bennett PM. 2004. Rifampicin resistance and its fitness cost in Enterococcus faecium. J. Antimicrob. Chemother. 53:203–207 [DOI] [PubMed] [Google Scholar]
- 28. Maharjan RP, Seeto S, Ferenci T. 2007. Divergence and redundancy of transport and metabolic rate-yield strategies in a single Escherichia coli population. J. Bacteriol. 189:2350–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kampmann M-L, Fordyce SL, Avila-Arcos MC, Rasmussen M, Willerslev E, Nielsen LP, Gilbert MTP. 2011. A simple method for the parallel deep sequencing of full influenza A genomes. J. Virol. Methods 178:243–248 [DOI] [PubMed] [Google Scholar]
- 30. Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247 [DOI] [PubMed] [Google Scholar]
- 31. Blattner F. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 32. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 35. Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. 2009. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics Chapter 13:Unit 13.11 [DOI] [PubMed] [Google Scholar]
- 36. Wimpenny JWT, Abdollahi H. 1991. Growth of mixed cultures of Paracoccus denitrificans and Desulfovibrio desulfuricans in homogeneous and in heterogeneous culture systems. Microb. Ecol. 22:1–13 [DOI] [PubMed] [Google Scholar]
- 37. Shehata T, Marr A. 1975. Effect of temperature on the size of Escherichia coli cells. J. Bacteriol. 124:857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. King T, Ishihama A, Kori A, Ferenci T. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186:5614–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hanko VP, Rohrer JS. 2004. Determination of amino acids in cell culture and fermentation broth media using anion-exchange chromatography with integrated pulsed amperometric detection. Anal. Biochem. 324:29–38 [DOI] [PubMed] [Google Scholar]
- 41. Kim BH, Gadd GM. 2008. Bacterial physiology and metabolism. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 42. Geanacopoulos M, Adhya S. 1997. Functional characterization of roles of GalR and GalS as regulators of the gal regulon. J. Bacteriol. 179:228–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang Z, Bielawski J. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elena SF, Lenski RE. 2003. Microbial genetics: evolution experiments with microorganisms. The dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457–469 [DOI] [PubMed] [Google Scholar]
- 45. Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. 2006. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 103:9107–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wichman HA, Yarber CD, Scott LA, Bull JJ. 2000. Experimental evolution recapitulates natural evolution. A remarkable study of parallel and convergent molecular evolution that is based on the whole-genome sequencing of viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pelosi L, Kuhn L, Guetta D, Garin J, Geiselmann J, Lenski RE, Schneider D. 2006. Parallel changes in global protein profiles during long-term experimental evolution in Escherichia coli. Genetics 173:1851–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adhya SL, Shapiro JA. 1969. The galactose operon of E. coli K-12. I. Structural and pleiotropic mutations of the operon. Genetics 62:231–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tokeson JPE, Garges S., Adhya S. 1991. Further inducibility of a constitutive system: ultrainduction of the gal operon. J. Bacteriol. 73:2319–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wee S, Wilkinson B. 1988. Increased outer membrane ornithine-containing lipid and lysozyme penetrability of Paracoccus denitrificans grown in a complex medium deficient in divalent cations. J. Bacteriol. 170:3283–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Habets M, Rozen DE, Hoekstra RF, DE Visser JA. 2006. The effect of population structure on the adaptive radiation of microbial populations evolving in spatially structured environments. Ecol. Lett. 9:1041–1048 [DOI] [PubMed] [Google Scholar]
- 52. Flores N, Leal L, Sigala JC, de Anda R, Escalante A, Martínez A, Ramírez O., Gosset G, Bolivar F. 2007. Growth recovery on glucose under aerobic conditions of an Escherichia coli strain carrying a phosphoenolpyruvate:carbohydrate phosphotransferase system deletion by inactivating arcA and overexpressing the genes coding for glucokinase and galactose permease. J. Mol. Microbiol. Biotechnol. 13:105–116 [DOI] [PubMed] [Google Scholar]
- 53. Blaby I. 2012. Experimental evolution of a facultative thermophile from a mesophilic ancestor. Appl. Environ. Microbiol. 78:144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, Palsson BØ. 2009. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 10:R118.1–R118.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kassen R. 2009. Toward a general theory of adaptive radiation. The year in evolutionary biology. Ann. N. Y. Acad. Sci. 1168:3–22 [DOI] [PubMed] [Google Scholar]
- 56. Schluter D. 2000. The ecology of adaptive radiation. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 57. Notley-McRobb L, Ferenci T. 1999. Adaptive mgl-regulatory mutations and genetic diversity evolving in glucose-limited E. coli populations. Environ. Microbiol. 1:33–43 [DOI] [PubMed] [Google Scholar]
- 58. Kurlandzka A, Rosenzweig RF, Adams J. 1991. Identification of adaptive changes in an evolving population of Escherichia coli: the role of changes with regulatory and highly pleiotropic effects. Mol. Biol. Evol. 8:261–281 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.