Abstract
Two groups independently sequenced the Agrobacterium tumefaciens C58 genome in 2001. We report here consolidation of these sequences, updated annotation, and additional analysis of the evolutionary history of the linear chromosome, which is apparently limited to the biovar I group of Agrobacterium.
TEXT
Agrobacterium tumefaciens C58 has an unusual genome structure consisting of one circular chromosome (chromosome I), one linear chromosome (chromosome II), and two plasmids (1–5). Previous studies showed that the linear chromosome is derived from a plasmid (4, 5). Isolates of Agrobacterium spp. have traditionally been subdivided into three different groups, called biovars, based on differences in physiology and host range. Biovar I can be further subdivided into genomovars, with C58 belonging to genomovar 8 (6–10).
C58 was originally isolated in 1958 by Robert Dickey from a cherry gall in upstate New York (11). Lead authors of this article independently sequenced the genomes of two isolates of A. tumefaciens C58 in 2001 (4, 5). Wood et al. (5) sequenced a C58 strain stored in frozen glycerol in the laboratory of Eugene Nester at the University of Washington (hereafter designated C58UW). Goodner et al. (4) sequenced the ATCC 33970 isolate obtained from the American Type Culture Collection (ATCC) in 1999. This strain, also originating from the Nester lab via John Kleyn, was deposited in 1981 and subcultured three times by ATCC and once by researchers at the Monsanto Company prior to sequencing. The number of passages separating these strains from each other or the original strain isolated by Dickey is unknown.
A comparison of the two independent genome sequences identified 52 differences, including two insertion/deletions (indels) (see Table S1 in the supplemental material). All disparate loci were resequenced following PCR amplification (See the supplemental Materials and Methods in the supplemental material). Twenty-two of these apparent differences were base-calling errors, and 30 were true differences. Of the 30 true differences, 16 were single base changes residing in the 16S rRNA and tRNA-Ile region near 58.3 kbp on chromosome I, apparently resulting from recombination between rRNA loci. C58UW also contains two deletions relative to ATCC 33970. The first is a 90-bp in-frame deletion within a putative two-component response regulator gene (atu5121). The second is a 111-bp symmetrical intergenic deletion on the circular chromosome that removes part of a short repeat sequence called CIR2 (12, 13).
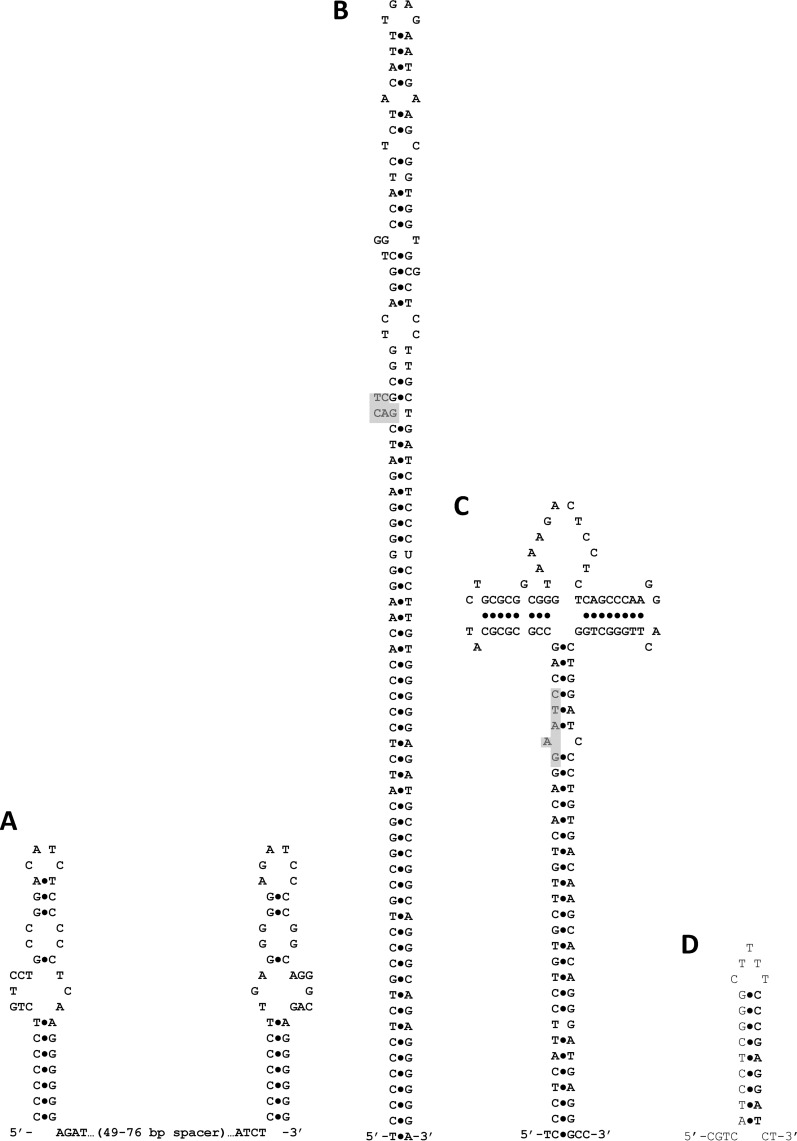
The latter result prompted a broader search for short repeated palindromic sequences within the C58 genome, resulting in the identification of three classes of repeats (Fig. 1). Two of these sequences, AgroCIR1 and AgroCIR2, were previously identified in a search for conserved motifs containing a binding site (GANTC) for the essential methylase CcrM (12, 13). The third element is herein designated AgroKE3 and bears no resemblance to the CIR repeats. A full KE3 repeat consists of 29-bp inverted repeats bracketing a variable region containing 49 to 76 bp (Fig. 1). Like AgroCIR1 and AgroCIR2, KE3 elements are preferentially found on chromosome I, consistent with the evolution of these repeats on the ancestral chromosome during the radiation of the Rhizobiaceae prior to the origins of chromosome II. Table S2 in the supplemental material summarizes the distribution of KE3 repeats in several closely related, fully sequenced relatives of A. tumefaciens C58, including the recently sequenced biovar I strain Agrobacterium sp. strain H13-3 (14). Table S3 in the supplemental material lists locations where these sequence repeats overlap a predicted open reading frame (ORF) in the C58 genome. The biological function of the KE3 repeats has not yet been determined.
Fig 1.
Potential secondary structures for genomic repeat elements. (A) The structure emphasizes the inverted repeats present at each end of the CIR2 full element. (B) An alternative secondary structure for a CIR2 full element that emphasizes the larger overall inverted repeat. The extent of the stem forming above position 44 depends on the sequence of the spacer region. The shaded polygon indicates where a CcrM binding site sits if one is present (it is found in only about one-third of the elements). (C) Potential secondary structure for CIR1 elements. The structure emphasizes the larger overall inverted repeat. The shaded area indicates the usual location of a CcrM binding site, but quite often, a second CcrM binding site exists directly opposite the first location in a full element (the example here does not contain such a second site). (D) Potential secondary structure for KE3 half elements and ends of KE3 full elements.
All true variant loci between C58UW and ATCC 33970 were compared to the same loci in other A. tumefaciens C58 culture lines obtained from laboratories in the United States and Europe (see Tables S1 and S4 in the supplemental material). In 12 of 14 cases, including both indels, the ATCC 33970 sequence was identical to each of the C58 comparison strains, while in two cases, all reference strains matched C58UW. While the cause of the additional variation in C58UW is unclear, it may be that the strain was passaged more frequently or that one of the acquired variations resulted in a higher mutation rate.
The telomeres of the C58 linear chromosome are covalently closed hairpin loops (4). This unusual structure meant that neither of our original studies was able to provide a complete sequence for its ends; similarly, the telomeres have not yet been sequenced for H13-3 (14). Recently, however, the C58 telomere sequences, along with a biochemical characterization of the protelomerase enzyme that maintains them (TelA, encoded by atu2523), have been published (15). The updated GenBank submission has been modified to include these data (see below).
We hypothesize that linearization of chromosome II was a seminal event in the divergence of biovar I strains, such as A. tumefaciens C58, from biovar III strains, such as Agrobacterium vitis S4 (16). The simplest model for its linearization involves a single crossover between the ancestral circular chromosome II and a linear phage or plasmid, thereby incorporating both telomeres and telA into the genome in one event. Surprisingly, however, the telA gene is located on the circular chromosome I (4, 16). Comparison of the C58 and S4 genomes shows significant synteny between a single region of S4 chromosome I and three regions of the C58 chromosome. Our analysis of these relationships suggests that multiple recombination events in the atu2521-atu2523 (telA) region transferred telA to chromosome I and initiated two large DNA transfers to chromosome II (Fig. 2). The breakpoint for the translocation of genes atu3507 through atu8188 (0.558 to 0.575 Mbp on chromosome I of C58) from the circular to the linear chromosome is adjacent to telA. A similar translocation breakpoint occurs on chromosome I immediately upstream of atu2521 and extends through atu4172 (lysC) into the adjacent rRNA loci (1.311 to 1.292 Mbp on chromosome I of C58). These genomic reorganizations transferred an rRNA operon and several essential genes to chromosome II while placing telA on chromosome I. Intriguingly, atu2521 and atu2522 are more similar to orthologs in Rhizobium and Sinorhizobium, respectively, than they are to their orthologs in S4 (avi3961 and avi3963, respectively), suggesting that atu2521, atu2522, and telA may have entered the C58 genome together, perhaps as part of a linear plasmid.
Fig 2.
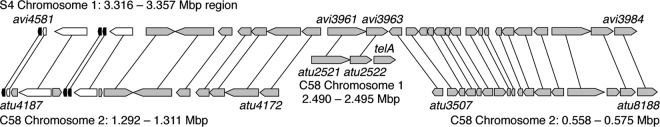
Genomic transfers between chromosome I and chromosome II associated with the atu2521-atu2523 (telA) region. The top diagram shows genes on the A. vitis S4 chromosome I from avi4581 (tRNA-met) through avi3984 (a conserved hypothetical gene) (16). The lower diagrams show the syntenic regions of the A. tumefaciens C58 genome, with lines connecting orthologous genes of C58 and S4. Note the breakpoints in synteny immediately upstream of avi3961 and downstream of avi3963 in the S4 genome, identifying breakpoints for large genomic transfers between chromosome I and chromosome II in the C58 genome. Protein-coding genes are shown in gray, tRNA genes are shown in black, and rRNA genes are shown in white. Genomic locations of each region correspond to the base pair numbering in the sequence files available at NCBI.
We surveyed a large number of strains that have historically been classified as Agrobacterium, including biovar I (A. tumefaciens), biovar II (Agrobacterium radiobacter), and biovar III (A. vitis), for the presence of a linear mega-size DNA molecule by pulsed-field gel electrophoresis (PFGE) and for telA and its adjacent ORF, atu2522 (acvB), by PCR or a Southern blot (see Table S5 and Fig. S1 in the supplemental material). It is important to note in considering this analysis that strong evidence supports the reclassification of biovar II strains as Rhizobium (6, 10, 16–19). Our survey data indicate that linear chromosomes are unique to biovar I strains (see Table S5 in the supplemental material) (14, 20). Based on this comparison, we can now define the unique genomic content of biovar I as containing a linear replicon accompanied by a telA gene, in addition to other diagnostic genes (see Table S6 in the supplemental material).
We have added the recently published telomere sequences and consolidated our two earlier versions of the C58 genome sequence into a single version with updated annotation from our own work and that of others. ATCC 33970 was chosen as the standard sequence because it is most similar to other reference strains analyzed (see Table S1 in the supplemental material). Notations indicating the variations found in the C58UW strain are included in this update. The gene identifiers (locus tags) referring to genes kept from the original annotations are the same as those defined by Wood et al. (format, atuXXXX) (5). Newly predicted protein-coding genes were given the locus tag pattern atu8XXX, as were a number of genes that were initially predicted only by Goodner et al. (4) or by analyses subsequent to the initial genome deposit (21). Newly predicted small RNA genes (22) were designated with the locus tag pattern atu9xxx. Details of the reannotation are provided in the supplemental Materials and Methods in the supplemental material.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the consolidated sequences are as follows: chromosome I, AE007869; chromosome II, AE007870; pTiC58, AE007871; and pAtC58, AE007872. These sequence files replace the two original versions of the A. tumefaciens C58 sequence files submitted by our research groups (4, 5).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation grants 0333297, 0603491, and 0736671, by a grant from the M. J. Murdock Charitable Trust life sciences program (2004262:JVZ), by a science education grant from the Howard Hughes Medical Institute (52005125), and by the Monsanto Fund.
We thank Kelly Williams for pointing out the group I intron within the tRNA gene. We thank the hundreds of students at Hiram College, Seattle Pacific University, the University of Arizona, and the Mesa (Arizona) Biotechnology Academy who have contributed to the annotation of this genome.
Footnotes
Published ahead of print 14 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03192-12.
REFERENCES
- 1. Allardet-Servent A, Michaux-Charachon S, Jumas-Bilak E, Karayan L, Ramuz M. 1993. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J. Bacteriol. 175:7869–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jumas-Bilak E, Michaux-Charachon S, Bourg G, Ramuz M, Allardet-Servent A. 1998. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J. Bacteriol. 180:2749–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goodner BW, Markelz BP, Flanagan MC, Crowell CB, Jr, Racette JL, Schilling BA, Halfon LM, Mellors JS, Grabowski G. 1999. Combined genetic and physical map of the complex genome of Agrobacterium tumefaciens. J. Bacteriol. 181:5160–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao YW, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slater S. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328 [DOI] [PubMed] [Google Scholar]
- 5. Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF, Woo L, Chen YC, Paulsen IT, Eisen JA, Karp PD, Bovee D, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu ZN, Romero P, Gordon D, Zhang SP, Yoo HY, Tao YM, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323 [DOI] [PubMed] [Google Scholar]
- 6. Portier P, Fischer-Le Saux M, Mougel C, Lerondelle C, Chapulliot D, Thioulouse J, Nesme X. 2006. Identification of genomic species in Agrobacterium biovar 1 by AFLP genomic markers. Appl. Environ. Microbiol. 72:7123–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lassalle F, Campillo T, Vial L, Baude J, Costechareyre D, Chapulliot D, Shams M, Abrouk D, Lavire C, Oger-Desfeux C, Hommais F, Gueguen L, Daubin V, Muller D, Nesme X. 2011. Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens. Genome Biol. Evol. 3:762–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costechareyre D, Bertolla F, Nesme X. 2009. Homologous recombination in Agrobacterium: potential implications for the genomic species concept in bacteria. Mol. Biol. Evol. 26:167–176 [DOI] [PubMed] [Google Scholar]
- 9. Mougel C, Thioulouse J, Perriere G, Nesme X. 2002. A mathematical method for determining genome divergence and species delineation using AFLP. Int. J. Syst. Evol. Microbiol. 52:573–586 [DOI] [PubMed] [Google Scholar]
- 10. Costechareyre D, Rhouma A, Lavire C, Portier P, Chapulliot D, Bertolla F, Boubaker A, Dessaux Y, Nesme X. 2010. Rapid and efficient identification of Agrobacterium species by recA allele analysis: Agrobacterium recA diversity. Microb. Ecol. 60:862–872 [DOI] [PubMed] [Google Scholar]
- 11. Hamilton RH, Fall MZ. 1971. The loss of tumor-initiating ability in Agrobacterium tumefaciens by incubation at high temperature. Experientia 27:229–230 [DOI] [PubMed] [Google Scholar]
- 12. Chen SL, Shapiro L. 2003. Identification of long intergenic repeat sequences associated with DNA methylation sites in Caulobacter crescentus and other α-proteobacteria. J. Bacteriol. 185:4997–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kahng LS, Shapiro L. 2001. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J. Bacteriol. 183:3065–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wibberg D, Blom J, Jaenicke S, Kollin F, Rupp O, Scharf B, Schneiker-Bekel S, Sczcepanowski R, Goesmann A, Setubal JC, Schmitt R, Puhler A, Schluter A. 2011. Complete genome sequencing of Agrobacterium sp. H13-3, the former Rhizobium lupini H13-3, reveals a tripartite genome consisting of a circular and a linear chromosome and an accessory plasmid but lacking a tumor-inducing Ti-plasmid. J. Biotechnol. 155:50–62 [DOI] [PubMed] [Google Scholar]
- 15. Huang WM, Dagloria J, Fox H, Ruan Q, Tillou J, Shi K, Aihara H, Aron J, Casjens S. 2012. Linear chromosome generating system of Agrobacterium tumefaciens C58: protelomerase generates and protects hairpin ends. J. Biol. Chem. 287:25551–25563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, Burr TJ, Banta L, Dickerman AW, Paulsen I, Otten L, Suen G, Welch R, Almeida NF, Arnold F, Burton OT, Du Z, Ewing A, Godsy E, Heisel S, Houmiel KL, Jhaveri J, Lu J, Miller NM, Norton S, Chen Q, Phoolcharoen W, Ohlin V, Ondrusek D, Pride N, Stricklin SL, Sun J, Wheeler C, Wilson L, Zhu H, Wood DW. 2009. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 191:2501–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Lajudie P, Laurent-Fulele E, Willems A, Torck U, Coopman R, Collins MD, Kersters K, Dreyfus B, Gillis M. 1998. Allorhizobium undicola gen. nov., sp. nov., nitrogen-fixing bacteria that efficiently nodulate Neptunia natans in Senegal. Int. J. Syst. Bacteriol. 48:1277–1290 [DOI] [PubMed] [Google Scholar]
- 18. Sawada H, Ieki H, Oyaizu H, Matsumoto S. 1993. Proposal for rejection of Agrobacterium tumefaciens and revised descriptions for the genus Agrobacterium and for Agrobacterium radiobacter and Agrobacterium rhizogenes. Int. J. Syst. Bacteriol. 43:694–702 [DOI] [PubMed] [Google Scholar]
- 19. Young JM, Kuykendall LD, Martínez-Romero E, Kerr A, Sawada H. 2001. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J. Syst. Evol. Microbiol. 51:89–103 [DOI] [PubMed] [Google Scholar]
- 20. Li A, Geng J, Cui D, Shu C, Zhang S, Yang J, Xing J, Wang J, Ma F, Hu S. 2011. Genome sequence of Agrobacterium tumefaciens strain F2, a bioflocculant-producing bacterium. J. Bacteriol. 193:5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Q, Lei Y, Xu X, Wang G, Chen LL. 2012. Theoretical prediction and experimental verification of protein-coding genes in plant pathogen genome Agrobacterium tumefaciens strain C58. PLoS One 7:e43176 doi:10.1371/journal.pone.0043176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilms I, Overloper A, Nowrousian M, Sharma CM, Narberhaus F. 2012. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol. 9:446–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.